Difference between revisions of "Part:BBa K1529300"
| Line 30: | Line 30: | ||
III. Comparison of the improved Prhl by iGEM 2014 Tokyo_Tech to our original improved Prhl | III. Comparison of the improved Prhl by iGEM 2014 Tokyo_Tech to our original improved Prhl | ||
| − | |||
| − | |||
| − | |||
<span style="margin-left: 10px;">Our purpose is to create strong Prhl for our final genetic circuits. | <span style="margin-left: 10px;">Our purpose is to create strong Prhl for our final genetic circuits. | ||
| − | |||
| − | + | The past improved Prhl did not suit for our final circuits and we could construct the improved Prhl appropriate to our final circuits. | |
| − | |||
| − | + | ====I. Improved Prhl by iGEM 2014 Tokyo_Tech team and characterization of <i>RhlR</i>==== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | ====I. Improved Prhl by iGEM 2014 Tokyo_Tech and | + | |
<span style="margin-left: 10px;">We found that Prhl(RL) (<partinfo>BBa_K1529300</partinfo>) activity was weak and the expression level depended on LVA tag (Fig.1); LVA-tagged proteins are prone to be degraded by cellular proteases. Prhl(LR) (<partinfo>BBa_K1529310</partinfo>) activity was strong and unexpectedly reacted with C12 (crosstalk) (Fig.3). | <span style="margin-left: 10px;">We found that Prhl(RL) (<partinfo>BBa_K1529300</partinfo>) activity was weak and the expression level depended on LVA tag (Fig.1); LVA-tagged proteins are prone to be degraded by cellular proteases. Prhl(LR) (<partinfo>BBa_K1529310</partinfo>) activity was strong and unexpectedly reacted with C12 (crosstalk) (Fig.3). | ||
| Line 55: | Line 45: | ||
[[Image:prhl2.png|thumb|center|400px|Fig.2 The colonies of transformants with a rhlR (left) or a rhlR-LVA (right)]]<br> | [[Image:prhl2.png|thumb|center|400px|Fig.2 The colonies of transformants with a rhlR (left) or a rhlR-LVA (right)]]<br> | ||
| − | |||
====II. Improvement the wild type Prhl==== | ====II. Improvement the wild type Prhl==== | ||
| Line 63: | Line 52: | ||
[[Image:prhl4.png|thumb|center|400px|Fig. 3 RFU of GFP / turbidity of imoroved Prhl mutants]]<br> | [[Image:prhl4.png|thumb|center|400px|Fig. 3 RFU of GFP / turbidity of imoroved Prhl mutants]]<br> | ||
| − | The sequences of these | + | The sequences of these two chosen mutants are shown below. The small characters indicate the scar sequence and a single point mutation is colored with red. |
| − | ====III. Comparison the improved Prhl by iGEM 2014 Tokyo_Tech to our original improved Prhl==== | + | Native Prhl (<partinfo>BBa_R0071</partinfo>) |
| + | |||
| + | TCCTGTGAAATCTGGCAGTTACCGTTAGCTTTCGAATTGGCTAAAAAGTGTTC | ||
| + | |||
| + | Prhl(NM) (<partinfo>BBa_K1949060</partinfo>) | ||
| + | |||
| + | TCCTGTGAAATCTGGCAGTTACCGTTAGCTTTCGAATTGGCTA<font color=#ff0000><b>t</b></font>AAAGTGTTC | ||
| + | |||
| + | |||
| + | ====III. Comparison of the improved Prhl by iGEM 2014 Tokyo_Tech team to our original improved Prhl==== | ||
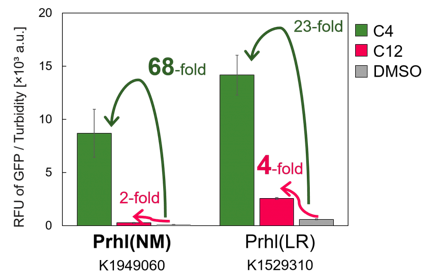
<span style="margin-left: 10px;">Prhl(NM) was chosen from the many Prhl mutants, and by comparing Prhl(NM) to Prhl(LR), we obtained the result below (Fig.4). The reaction activity of Prhl(NM) to C4 was stronger than that of Prhl(LR), and Prhl(NM) did not react with C12 at all. | <span style="margin-left: 10px;">Prhl(NM) was chosen from the many Prhl mutants, and by comparing Prhl(NM) to Prhl(LR), we obtained the result below (Fig.4). The reaction activity of Prhl(NM) to C4 was stronger than that of Prhl(LR), and Prhl(NM) did not react with C12 at all. | ||
| − | [[Image: | + | [[Image:prhl.png|thumb|center|400px|Fig.4 Comparison of Prhl(NM) to Prhl(LR)]]<br> |
| + | |||
If you want more information, you see [http://2016.igem.org/Team:Tokyo_Tech our work in Tokyo_Tech 2016 wiki]! | If you want more information, you see [http://2016.igem.org/Team:Tokyo_Tech our work in Tokyo_Tech 2016 wiki]! | ||
Revision as of 20:16, 19 October 2016
Prhl(RL)
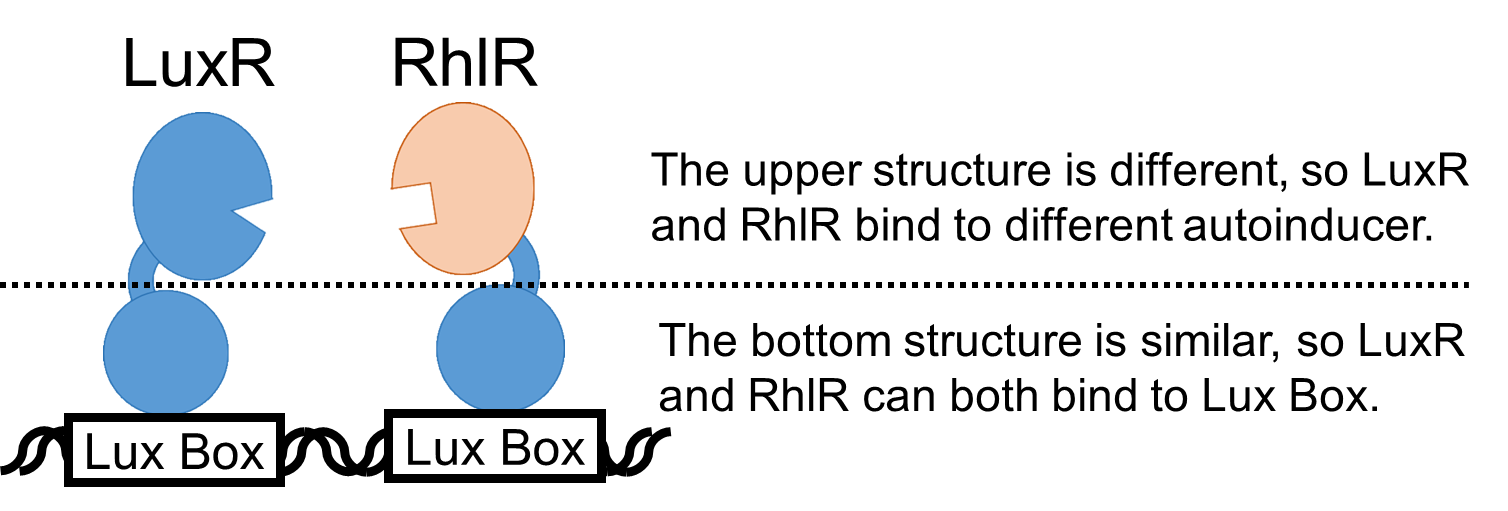
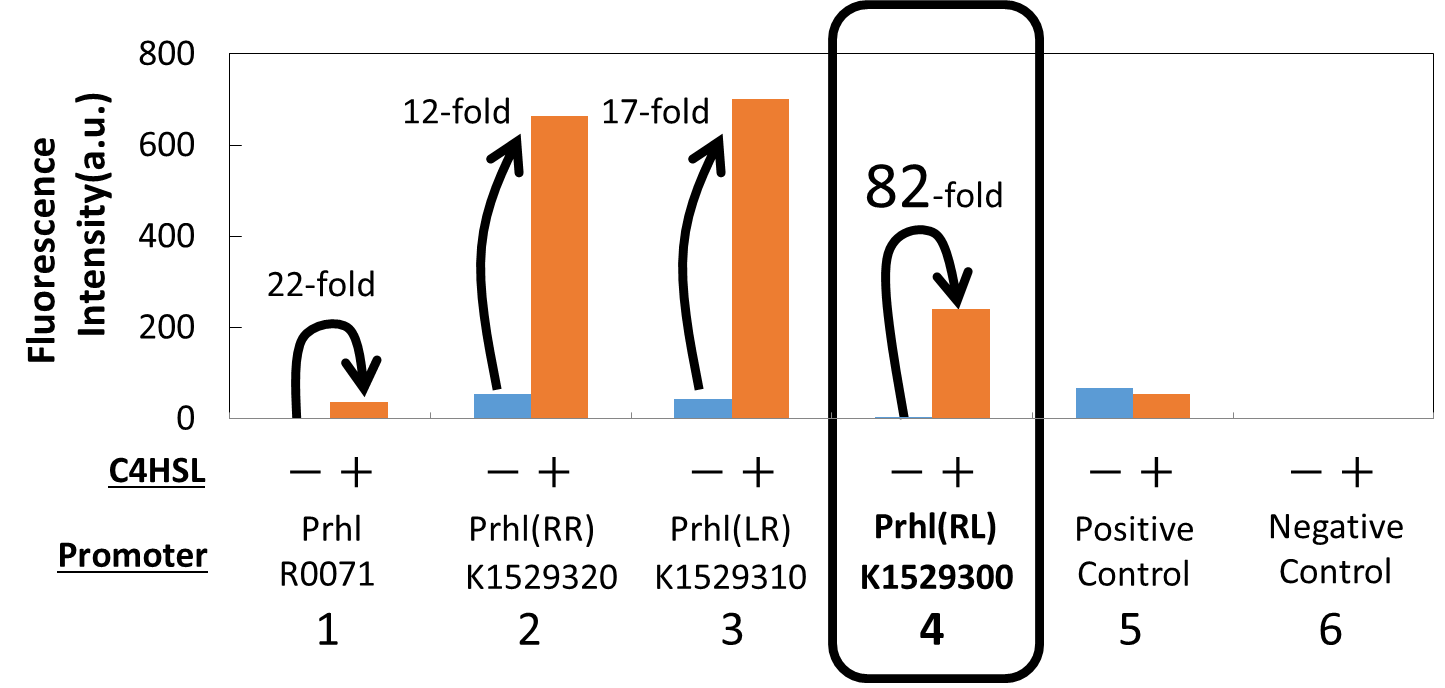
We newly developed the Prhl(RL) promoter (BBa_K1529300) by improving Prhl promoter (BBa_R0071) which is activated by C4HSL-RhlR complex (Fig. 1).
To characterize the function of this Prhl(RL) promoter, we constructed a part, Prhl(RL)-GFP (BBa_K1529301), by inserting the Prhl(RL) promoter, upstream of a GFP coding sequence.
By using the reporter cells that contain Prhl(RL)-GFP, we measured the fluorescence intensity of the cells induced by C4HSL. (Fig. 3.)
We saw that our new Prhl(RL) promoter was actually activated by C4HSL through an induction assay
For more information, see [http://2014.igem.org/Team:Tokyo_Tech/Experiment/Prhl_reporter_assay our work in Tokyo_Tech 2014 wiki].
Improvement and Characterization
Group: Tokyo Tech 2016
Author: Yoshio Takata
Summary of Improvement and Characterization:
I. Improved Prhl by iGEM 2014 Tokyo_Tech and characterize RhlR assay
II. Improvement of the wild type Prhl
III. Comparison of the improved Prhl by iGEM 2014 Tokyo_Tech to our original improved Prhl
Our purpose is to create strong Prhl for our final genetic circuits.
The past improved Prhl did not suit for our final circuits and we could construct the improved Prhl appropriate to our final circuits.
I. Improved Prhl by iGEM 2014 Tokyo_Tech team and characterization of RhlR
We found that Prhl(RL) (BBa_K1529300) activity was weak and the expression level depended on LVA tag (Fig.1); LVA-tagged proteins are prone to be degraded by cellular proteases. Prhl(LR) (BBa_K1529310) activity was strong and unexpectedly reacted with C12 (crosstalk) (Fig.3).
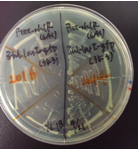
The colonies of transformants with a rhlR (BBa_C0171) plasmid looked rough and the growth rate was low(Fig.2-left), while the colonies of transformants with a rhlR-LVA (BBa_C0071) plasmid looked smooth and the growth rate was normal(Fig.2-right). However, the reason for this result is unclear.
II. Improvement the wild type Prhl
By introducing a single point mutation into wild type Prhl (BBa_R0071) by PCR, we obtained 198 Prhl mutants and chose the two Prhl mutants of which promoter activity was stronger than wild type Prhl(Fig.3).
The sequences of these two chosen mutants are shown below. The small characters indicate the scar sequence and a single point mutation is colored with red.
Native Prhl (BBa_R0071)
TCCTGTGAAATCTGGCAGTTACCGTTAGCTTTCGAATTGGCTAAAAAGTGTTC
Prhl(NM) (BBa_K1949060)
TCCTGTGAAATCTGGCAGTTACCGTTAGCTTTCGAATTGGCTAtAAAGTGTTC
III. Comparison of the improved Prhl by iGEM 2014 Tokyo_Tech team to our original improved Prhl
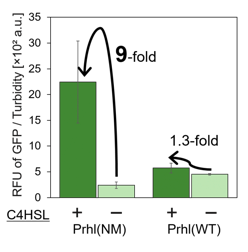
Prhl(NM) was chosen from the many Prhl mutants, and by comparing Prhl(NM) to Prhl(LR), we obtained the result below (Fig.4). The reaction activity of Prhl(NM) to C4 was stronger than that of Prhl(LR), and Prhl(NM) did not react with C12 at all.
If you want more information, you see [http://2016.igem.org/Team:Tokyo_Tech our work in Tokyo_Tech 2016 wiki]!
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]