Difference between revisions of "Part:BBa K1467101"
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1467101 short</partinfo> | <partinfo>BBa_K1467101 short</partinfo> | ||
| − | |||
35S is a plant specific promoter obtained from the Cauliflower Mosaic Virus. The part is intended for use as a constitutive promoter for gene expression experiments in plants. This part is compatible with GoldenGate MoClo Assembly Standard as it is free from internal BsaI and BpiI recognition sequences. | 35S is a plant specific promoter obtained from the Cauliflower Mosaic Virus. The part is intended for use as a constitutive promoter for gene expression experiments in plants. This part is compatible with GoldenGate MoClo Assembly Standard as it is free from internal BsaI and BpiI recognition sequences. | ||
| Line 23: | Line 22: | ||
<br> | <br> | ||
| − | The part is similar to parts BBa_K509000 and BBa_K414002 submitted by previous iGEM teams but has been domesticated to fit the GoldenGate MoClo Assembly Standard. The part can be used by future iGEM teams using GoldenGate | + | The part is similar to parts BBa_K509000 and BBa_K414002 submitted by previous iGEM teams but has been domesticated to fit the GoldenGate MoClo Assembly Standard. The part can be used by future iGEM teams using the GoldenGate standard, allowing cloning in a one-pot, one-step, digestion-ligation reaction as detailed by the NRP-UEA iGEM 2014 team protocols. |
Revision as of 16:28, 9 October 2014
GoldenGate compatible 35s Promoter
35S is a plant specific promoter obtained from the Cauliflower Mosaic Virus. The part is intended for use as a constitutive promoter for gene expression experiments in plants. This part is compatible with GoldenGate MoClo Assembly Standard as it is free from internal BsaI and BpiI recognition sequences.
iGEM14_NRP-UEA-Norwich have used this part in constructs to constitutively express proteins of interest, often providing a control to test the expression of each of our reporters. This allowed us to compare expression levels in contrast to when using promoters induced by external stimuli (BS3, PDF1.2, PR1), allowing us to select the most effective inducible promoters. Examples of the way in which the 35s promoter was used in The Green Canary project are shown below.
Figure 1: The result of an infiltration of Agrobacterium tumefaciens into Nicotiana benthamiana, showing plant cell apoptosis caused by expression of Bax. This Bax expression also gives evidence for the function of the 35S promoter which is constitutive and therefore causes expression of the protein without external stimuli.
Image (A) provides a control, showing a Nicotiana benthamiana leaf infiltrated (red circles) with Agrobacterium tumefaciens whilst B shows a Nicotiana benthamiana leaf infiltrated (red circles) with Agrobacterium tumefaciens carrying the plasmid encoding: 35s Constututive Promotor (Cauliflower Mosaic Virus) + Bax Coding Sequence (Mus Musculus) + Octopine Synthase Terminator (Agrobacterium tumefaciens).
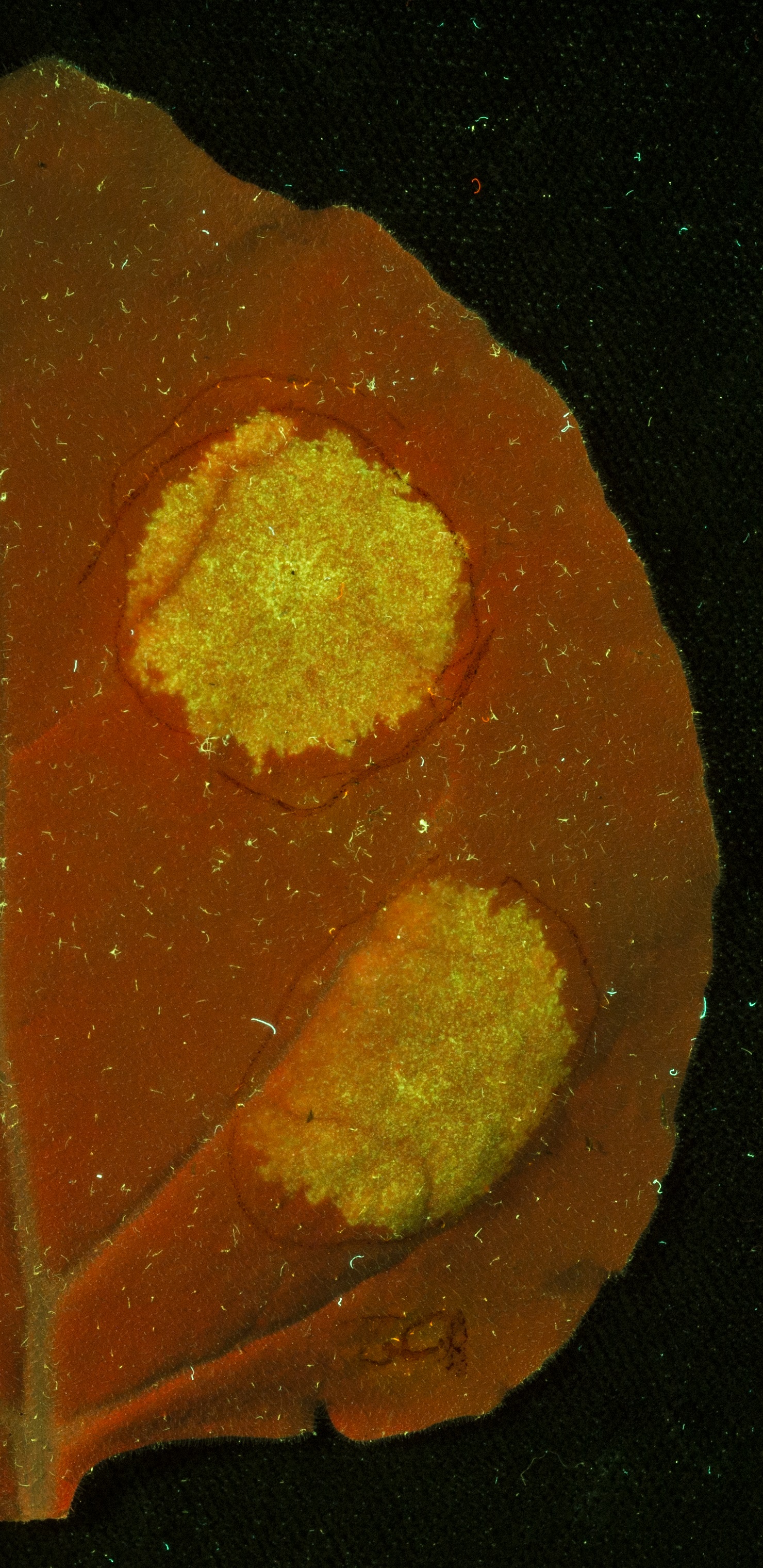
Figure 2: A Nicotiana benthamiana leaf under UV light following infiltration (red circles) with Agrobacterium tumefaciens carrying the plasmid encoding 35s Constututive Promotor (Cauliflower Mosaic Virus) + GFP Coding Sequence + Octopine Synthase Terminator (Agrobacterium tumefaciens). The result shows GFP expression, driven by the constitutive 35s promoter.
The part is similar to parts BBa_K509000 and BBa_K414002 submitted by previous iGEM teams but has been domesticated to fit the GoldenGate MoClo Assembly Standard. The part can be used by future iGEM teams using the GoldenGate standard, allowing cloning in a one-pot, one-step, digestion-ligation reaction as detailed by the NRP-UEA iGEM 2014 team protocols.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 392
Illegal BglII site found at 1138
Illegal XhoI site found at 1342 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]