Difference between revisions of "Part:BBa K1351031"
| Line 7: | Line 7: | ||
5' - GAATTC GCGGCCGC T TCTAGA G aggagaggata T ACTAGT A GCGGCCG CTGCAG - 3' | 5' - GAATTC GCGGCCGC T TCTAGA G aggagaggata T ACTAGT A GCGGCCG CTGCAG - 3' | ||
EcoRI NotI XbaI SpeI NotI PstI | EcoRI NotI XbaI SpeI NotI PstI | ||
| + | |||
| + | This part is used in the 2014 LMU-Munich iGEM project [http://2014.igem.org/Team:LMU-Munich BaKillus]. | ||
| + | |||
| + | This library was generated utilizing the [https://salis.psu.edu/software/ Salis lab RBS calculator] to find RBS that cover a high space of translation intitiation rate in 'B. subtilis'. It includes seven RBS with the following sequences: | ||
| + | |||
| + | {| class=wikitable | ||
| + | |- | ||
| + | !Number | ||
| + | !BBa_K1310 .. | ||
| + | !Sequence | ||
| + | |- | ||
| + | |1 | ||
| + | |28 | ||
| + | |aaggagggata | ||
| + | |- | ||
| + | |2 | ||
| + | |29 | ||
| + | |agaggaggata | ||
| + | |- | ||
| + | |<b>3</b> | ||
| + | |<b>30</b> | ||
| + | |<b>aaggagagata</b> | ||
| + | |- | ||
| + | |4 | ||
| + | |31 | ||
| + | |aggagaggata | ||
| + | |- | ||
| + | |5 | ||
| + | |32 | ||
| + | |aagaggagata | ||
| + | |- | ||
| + | |6 | ||
| + | |33 | ||
| + | |agaaagggata | ||
| + | |- | ||
| + | |7 | ||
| + | |34 | ||
| + | |aagaagagata | ||
| + | |} | ||
| + | |||
| + | The coverage of RBS strength as calculated by the [https://salis.psu.edu/software/ RBS calculator] is depicted in the Figure 1 shown below. It shows an evenly distributed strength spanning approxiamately three orders of magnitude. | ||
| + | |||
| + | [[File:LMU_14_RBSlibrary.png|thumb|center|400px|Fig.1: Coverage of the translation initiation rate of the RBS library as calculated by the [https://salis.psu.edu/software/ RBS calculator]]] | ||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
Revision as of 06:36, 1 November 2014
LMU Bacillus RBS collection 4
This part was generated in RFC10, and has the following prefix and suffix:
Prefix Suffix
5' - GAATTC GCGGCCGC T TCTAGA G aggagaggata T ACTAGT A GCGGCCG CTGCAG - 3'
EcoRI NotI XbaI SpeI NotI PstI
This part is used in the 2014 LMU-Munich iGEM project [http://2014.igem.org/Team:LMU-Munich BaKillus].
This library was generated utilizing the Salis lab RBS calculator to find RBS that cover a high space of translation intitiation rate in 'B. subtilis'. It includes seven RBS with the following sequences:
| Number | BBa_K1310 .. | Sequence |
|---|---|---|
| 1 | 28 | aaggagggata |
| 2 | 29 | agaggaggata |
| 3 | 30 | aaggagagata |
| 4 | 31 | aggagaggata |
| 5 | 32 | aagaggagata |
| 6 | 33 | agaaagggata |
| 7 | 34 | aagaagagata |
The coverage of RBS strength as calculated by the RBS calculator is depicted in the Figure 1 shown below. It shows an evenly distributed strength spanning approxiamately three orders of magnitude.
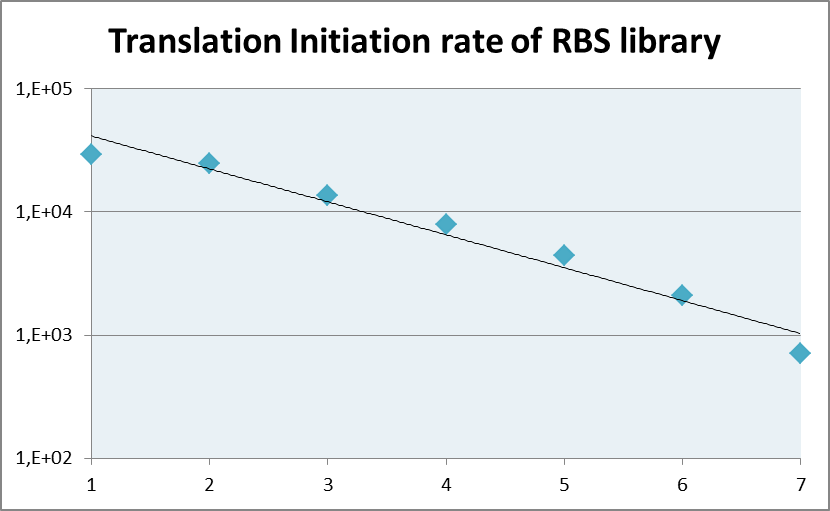
Fig.1: Coverage of the translation initiation rate of the RBS library as calculated by the RBS calculator
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
