Difference between revisions of "Part:BBa K1321014"
| Line 39: | Line 39: | ||
http://2014.igem.org/Team:Imperial/Water_Filtration | http://2014.igem.org/Team:Imperial/Water_Filtration | ||
| − | [[File:IC14_first_wash_CBD_metal_conc.png|700px| | + | [[File:IC14_first_wash_CBD_metal_conc.png|700px|centre|]] |
| − | [[File:IC14_second_wash_CBD_metal_conc.png|700px| | + | [[File:IC14_second_wash_CBD_metal_conc.png|700px|centre|]] |
| − | [[File:IC14_Comparison_Nickel_conc_washes_CBDs.png|700px| | + | [[File:IC14_Comparison_Nickel_conc_washes_CBDs.png|700px|centre|]] |
| − | [[File:IC-2014_metalbindingtable.jpg|900px| | + | [[File:IC-2014_metalbindingtable.jpg|900px|centre|]] |
Revision as of 04:50, 2 November 2014
CBDCipA with N and C-terminal linker
This CBD is from the Cellulosomal -scaffolding protein A (cipA) of Clostridium thermocellum including the endogenous linker sequences at the N and C-terminus (UniProt ID Q06851 link http://www.uniprot.org/uniprot/Q06851). It is in RFC25 format to allow for easy use in protein fusions.
The CBD is part of the CBM3 family (link to cazy http://www.cazy.org/CBM3.html). This CBD has been used in many application: in fusions with cell adhesion peptides to enhance the properties of cellulose as a cell-growth matrix (Andrade et al 2010a, Andrade et al 2010b), fused to enzymes to remove contaminants from water (Kauffmann et al 2000) and fused to an antimicrobial peptide (Ramos, Domingues & Gama 2010).
At present the cloning for these constructs is still in progress to correct an illegal EcoRI site which was identified in the parts with this CBD. This can be achieved with a silent mutation via site-directed mutagenesis and we aim to send these parts to the registry once this is complete.
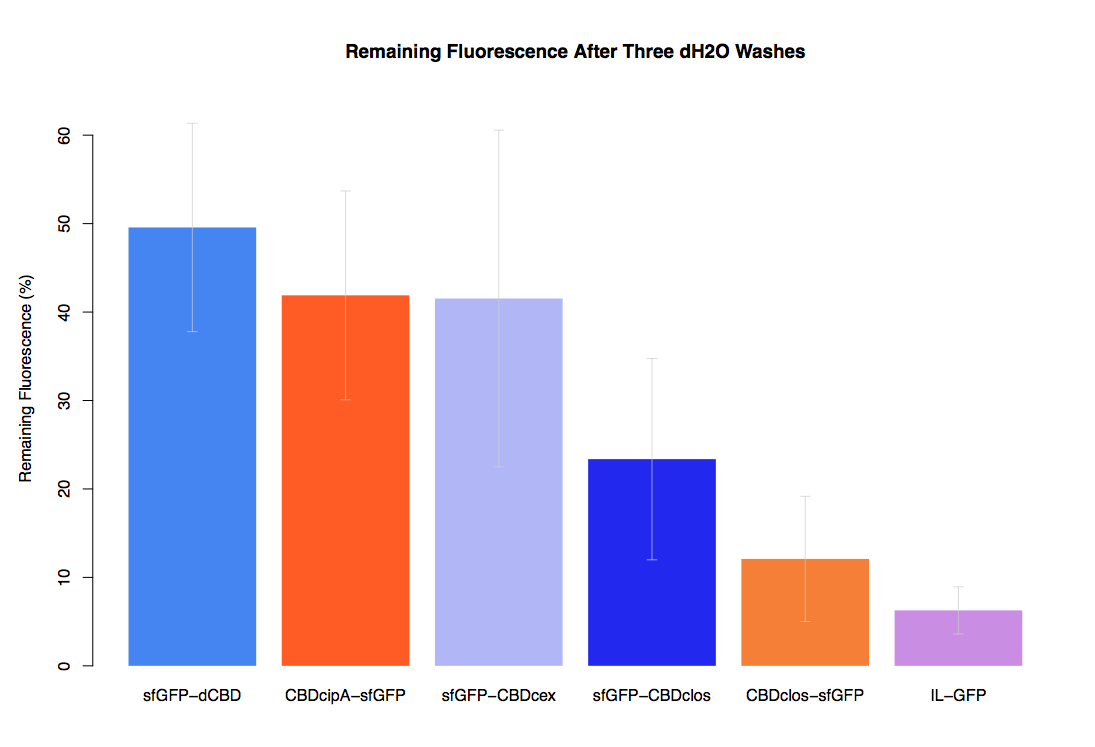
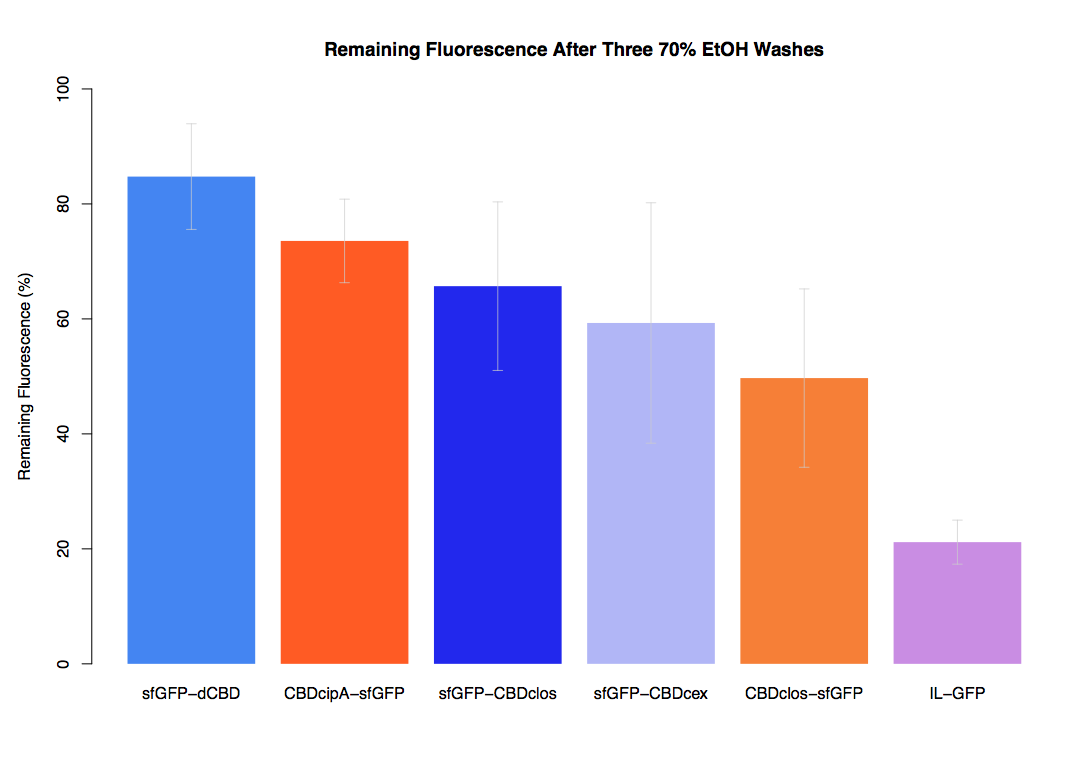
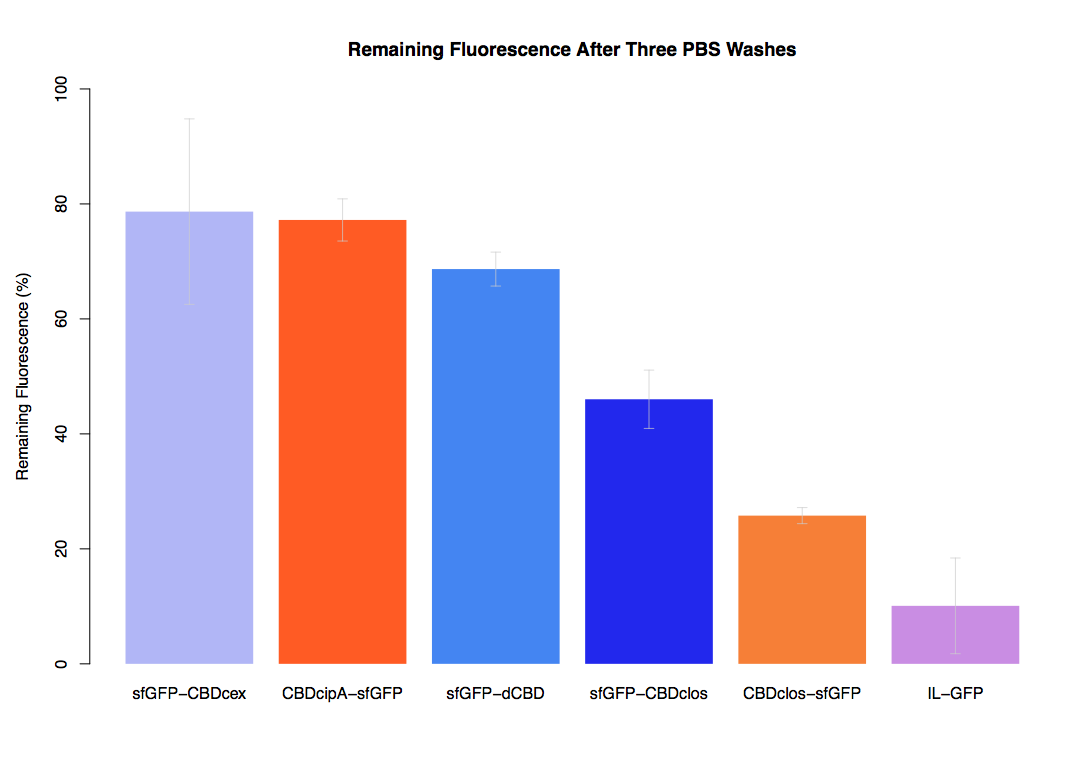
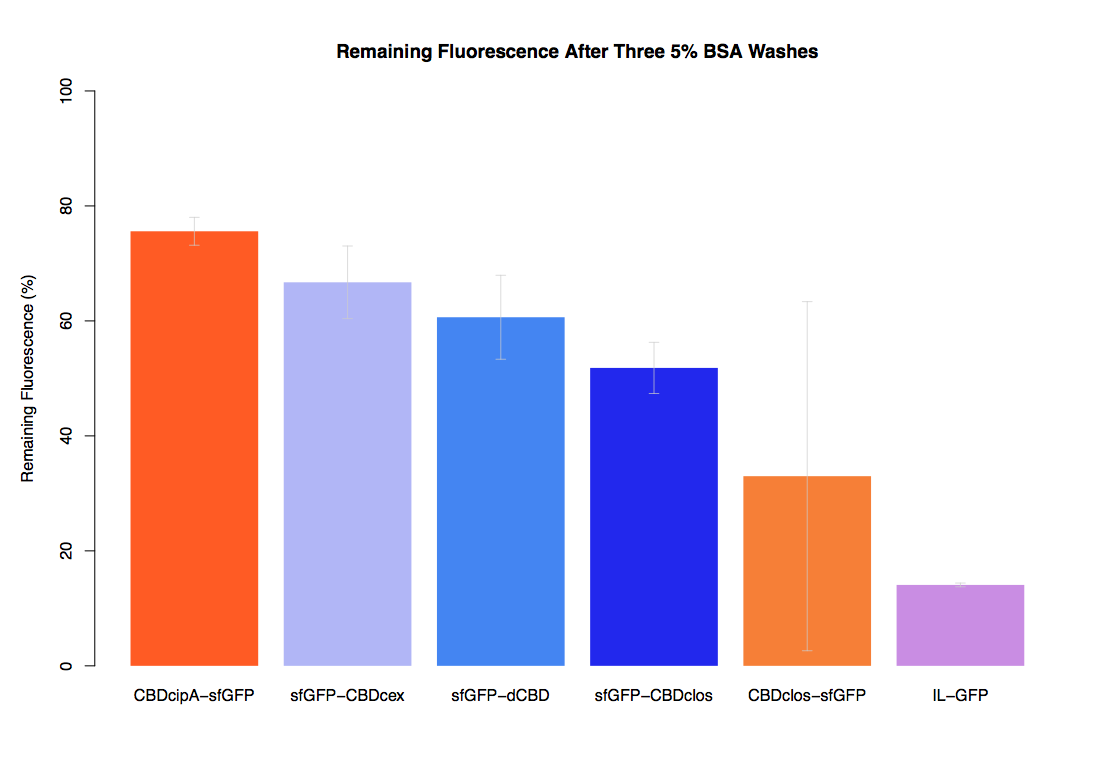
In our second assay performed to determine the relative strengths of various CBDs’ binding to bacterial cellulose – represented by the percentage fluorescence left from CBDs fused to sfGFP (RFC25) bound to bacterial cellulose discs, when subjected to various washes (protocol [http://2014.igem.org/Team:Imperial/Protocols here]) – it was determined that the CBDcipA-sfGFP fusion had the greatest binding ability in comparison to the four other CBDs fused to sfGFP after three washes with 5% BSA.
In the same assay, results suggested that CBDcipA that on average overall it had the second best ability to bind bacterial cellulose, as shown by the washes with PBS, 70% EtOH and dH2O.
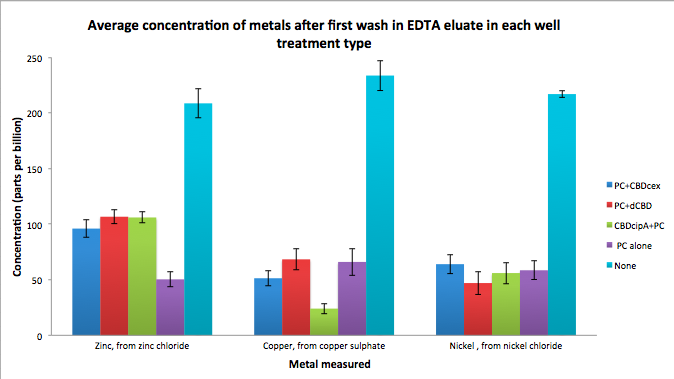
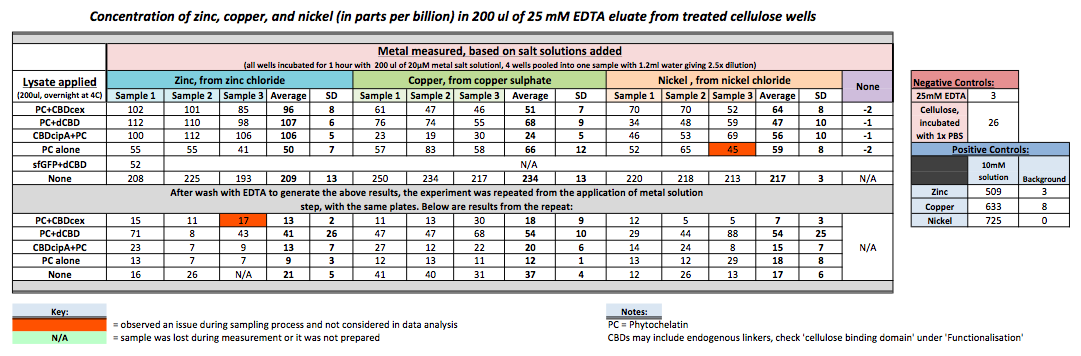
As part of our project, we needed to assay the metal binding capability of Phytochelatin (a general metal binding protein) fused to different CBDs (cellulose binding domains). The parts used for this assay are Phytochelatin+CBDcex, Phytochelatin+dCBD, CBDcipA+Phytochelatin, Phytochelatin alone and sfGFP+dCBD wash (only one wash).
Phytochelatin fused to our CBDs were bound onto cellulose that were dried in the bottom of 96-well plates and tested against 3 different metals (nickel, copper, zinc). First, the fusion protein cell lysate was incubated overnight in the cellulose wells. Following this, the metal salt solutions are added in excess into the wells. Finally, an EDTA step removes the bound metal ions into solution, and the metal concentration in solution is quantified by mass spectrometer. Multiple washes with PBS and water were done between each binding step, ensuring that the metal ions that are measured were released from the phytochelatin.
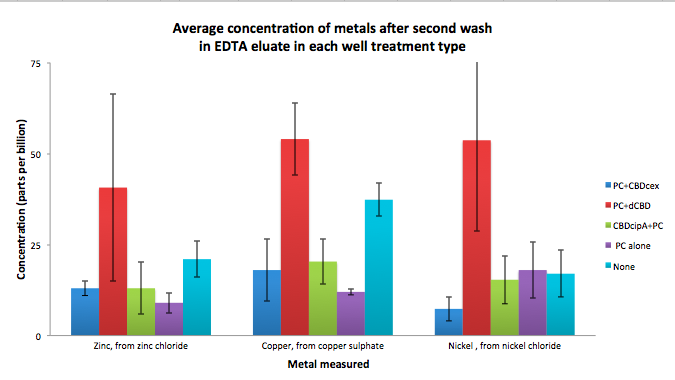
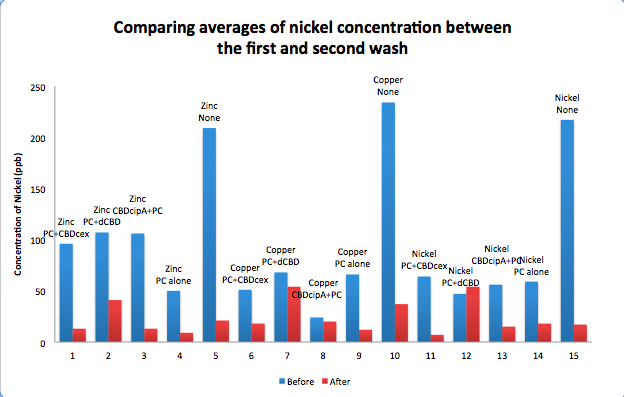
To evaluate if these CBD fusions were reusable, we re-applied metal ion solutions onto the same wells with the CBD fusions adhered. We washed the wells as before, then eluted with EDTA in the same manner. The results from the each elutions are shown below, with the final graph comparing between the first and second wash. We found along the way our bacterial cellulose alone had some metal chelating properties, as was also confirmed by our filtering set up with the dCBD-phytochelatin, but the second elution seems to show less background noise as the EDTA disrupts cellulose's natural binding to the metal ions. The full table of results are shown below as well. The full assay protocol can be found as ‘Metal binding assay protocol’ at this link: http://2014.igem.org/Team:Imperial/Protocols
To read more about this assay and explanation of these results, please see:
http://2014.igem.org/Team:Imperial/Water_Filtration
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 148
- 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 148
- 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 148
- 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 148
- 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 148
- 1000COMPATIBLE WITH RFC[1000]
References
Andrade, F.K., Moreira, S.M.G., Domingues, L. & Gama, F.M.P. (2010) Improving the affinity of fibroblasts for bacterial cellulose using carbohydrate-binding modules fused to RGD. Journal of biomedical materials research. Part A. [Online] 92 (1), 9–17. Available from: doi:10.1002/jbm.a.32284.
Andrade, F.K., Costa, R., Domingues, L., Soares, R., et al. (2010) Improving bacterial cellulose for blood vessel replacement: Functionalization with a chimeric protein containing a cellulose-binding module and an adhesion peptide. Acta biomaterialia. [Online] 6 (10), 4034–4041. Available from: doi:10.1016/j.actbio.2010.04.023.
Kauffmann, C., Shoseyov, O., Shpigel, E., Bayer, E.A., et al. (2000) Novel Methodology for Enzymatic Removal of Atrazine from Water by CBD-Fusion Protein Immobilized on Cellulose. Environmental Science & Technology. [Online] 34 (7), 1292–1296. Available from: doi:10.1021/es990754h.
Ramos, R., Domingues, L. & Gama, M. (2010) Escherichia coli expression and purification of LL37 fused to a family III carbohydrate-binding module from Clostridium thermocellum. Protein expression and purification. [Online] 71 (1), 1–7. Available from: doi:10.1016/j.pep.2009.10.016.