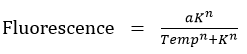Difference between revisions of "Part:BBa K1184000"
Chilam Poon (Talk | contribs) |
Chilam Poon (Talk | contribs) |
||
| Line 12: | Line 12: | ||
===Characterization=== | ===Characterization=== | ||
| − | ''' | + | '''2016 NYMU-Taipei team characterized this part with thermal stability assay.''' |
KillerRed protein was extracted from transformed E.coli and a control group of empty vector was set to eliminate the influence of other proteins. | KillerRed protein was extracted from transformed E.coli and a control group of empty vector was set to eliminate the influence of other proteins. | ||
| Line 55: | Line 55: | ||
| − | '''NYMU 2016 also developed this part to | + | '''NYMU 2016 also developed this part to construct a fungal expression cassette. ([https://parts.igem.org/Part:BBa_K2040121 BBa_K2040121])''' |
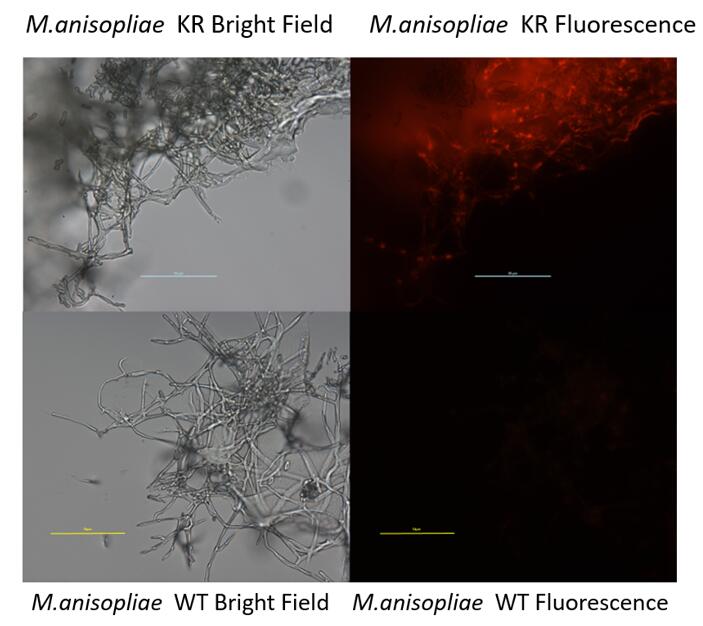
[[Image:PKT FL.jpeg|center|750px|thumb|Figure3. The bright field and fluorescence image of WT and KR transformants under Fluorescence Microscopy Olympus BX61]] | [[Image:PKT FL.jpeg|center|750px|thumb|Figure3. The bright field and fluorescence image of WT and KR transformants under Fluorescence Microscopy Olympus BX61]] | ||
Revision as of 02:37, 16 January 2017
KillerRed
KillerRed is a red fluorescent protein that produces reactive oxygen species (ROS) in the presence of yellow-orange light (540-585 nm). KillerRed is engineered from anm2CP to be phototoxic. It has been shown that KillerRed produces superoxide radical anions by reacting with water. Superoxide reacts with the chromophore of KillerRed, causing it to become dark, which ultimately gives rise to a bleaching effect. KillerRed is spectrally similar to mRFP1 with a similar brightness. KillerRed is oligomeric and may form large aggregates in cells. Expression of KillerRed and irradiation with light may act a kill-switch for biosafety applications.
Superoxide (O2•-) is the radical anion of molecular oxygen, which can react with protein side chains and lipids to harm cells.
This sequence is codon optimized for mammalian cells and has 13 rare proline codons for E. coli (CCC) and one rare arginine codon (AGA). Codon optimization should be taken into consideration if large amounts of the protein are required. Expression on a high-copy plasmid has produced detectable fluorescent signal, however.
Characterization
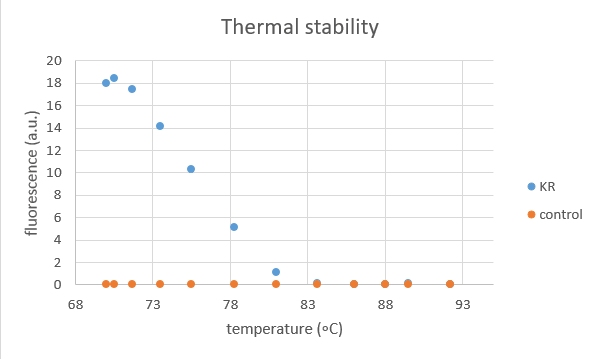
2016 NYMU-Taipei team characterized this part with thermal stability assay.
KillerRed protein was extracted from transformed E.coli and a control group of empty vector was set to eliminate the influence of other proteins.
This test aims to show the temperature influence on KillerRed protein by measuring its fluorescence after being treated for 2 hours at the following temperatures: 70, 70.5, 71.7, 73.5, 75.5, 78.3, 81, 83.6, 86, 88, 89.5, 92.2 (degree Celsius).
- It is obvious that KR contribute to most of the fluorescence.
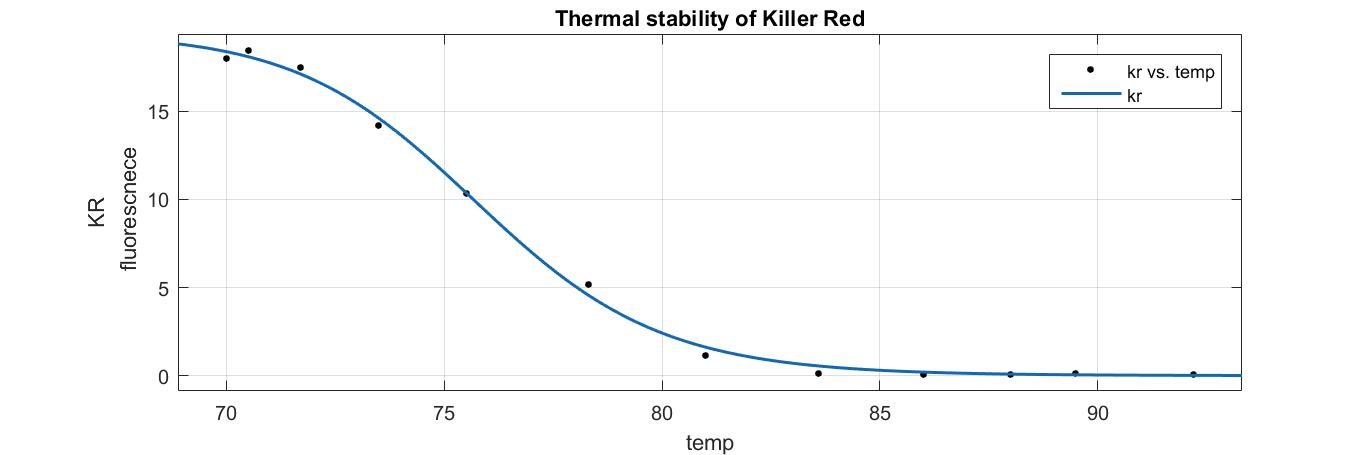
- A sigmoidal curve was fitted to find the particular temperature K at which half of the KR protein is denatured studied by measuring the fluorescence.
a=19.42
n=36.1
K=75.79
R-square=0.998
NYMU 2016 also developed this part to construct a fungal expression cassette. (BBa_K2040121)
- This new device was successfully expressed in M.anisopliae
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 151
Illegal BsaI.rc site found at 442