Difference between revisions of "Part:BBa K1159001"
CMichielsen (Talk | contribs) |
CMichielsen (Talk | contribs) |
||
| Line 42: | Line 42: | ||
<br> | <br> | ||
<h2> <b>Improved by TU_Eindhoven 2019</b> </h2> | <h2> <b>Improved by TU_Eindhoven 2019</b> </h2> | ||
| − | We developed a cysteine-free version of NanoLuc ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K3168006 BBa_K3168006]). By mutating the two cysteines to a serine, the luciferase can be used in fusion proteins that need to be labelled through maleimide coupling, such as our own dCas9-BRET sensor protein ([https://parts.igem.org/wiki/index.php?title=Part: | + | We developed a cysteine-free version of NanoLuc ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K3168006 BBa_K3168006]). By mutating the two cysteines to a serine, the luciferase can be used in fusion proteins that need to be labelled through maleimide coupling, such as our own dCas9-BRET sensor protein ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K3168007 BBa_K3168007]). |
<br> | <br> | ||
Revision as of 08:37, 15 October 2019
NanoLuc Luciferase in RFC[25]
NanoLuc Luciferase is engineered ATP-independent luciferase from a deep-sea shrimp which luminescense 2 magnitues higher than these from Renilla reniformis or from Photinus pyralis (firefly). Also the molecular weight of NanoLuc luciferase is twice smaller compared to other luciferase (only 19 kDa).
Usage and Biology
The Nano Luciferase (NanoLuc) which was introduced in 2013 by Promega is a new member of the luciferase reporter gene/protein familiy and shows some advantages compared to the other family members. The NanoLuc is very small (19 kDa) compared to the firefly luciferase (61 kDa) and the Renilla luciferase (36 kDa). On the other hand it is also said that the specific activity of the NanoLuc is about 150-fold stronger compared to conventional luciferases and the background caused by autoluminescense of the substrate is said to be remarkably smaller.
Protein Data Table for the Nano Luciferase BBa_K1159001
| Protein data table for BioBrick BBa_K1159001 automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCGTGTTCACC ... ATCCTTGCTACCGGT ORF from nucleotide position -8 to 516 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
Alignments (obtained from PredictProtein.org)
| ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Localization (reliability in brackets)
| Gene Ontology (reliability in brackets)
| |||||||||||||||||||||||||||||||||||||||||||||
Predicted features:
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
Experimental Data
Production in E. coli and purification
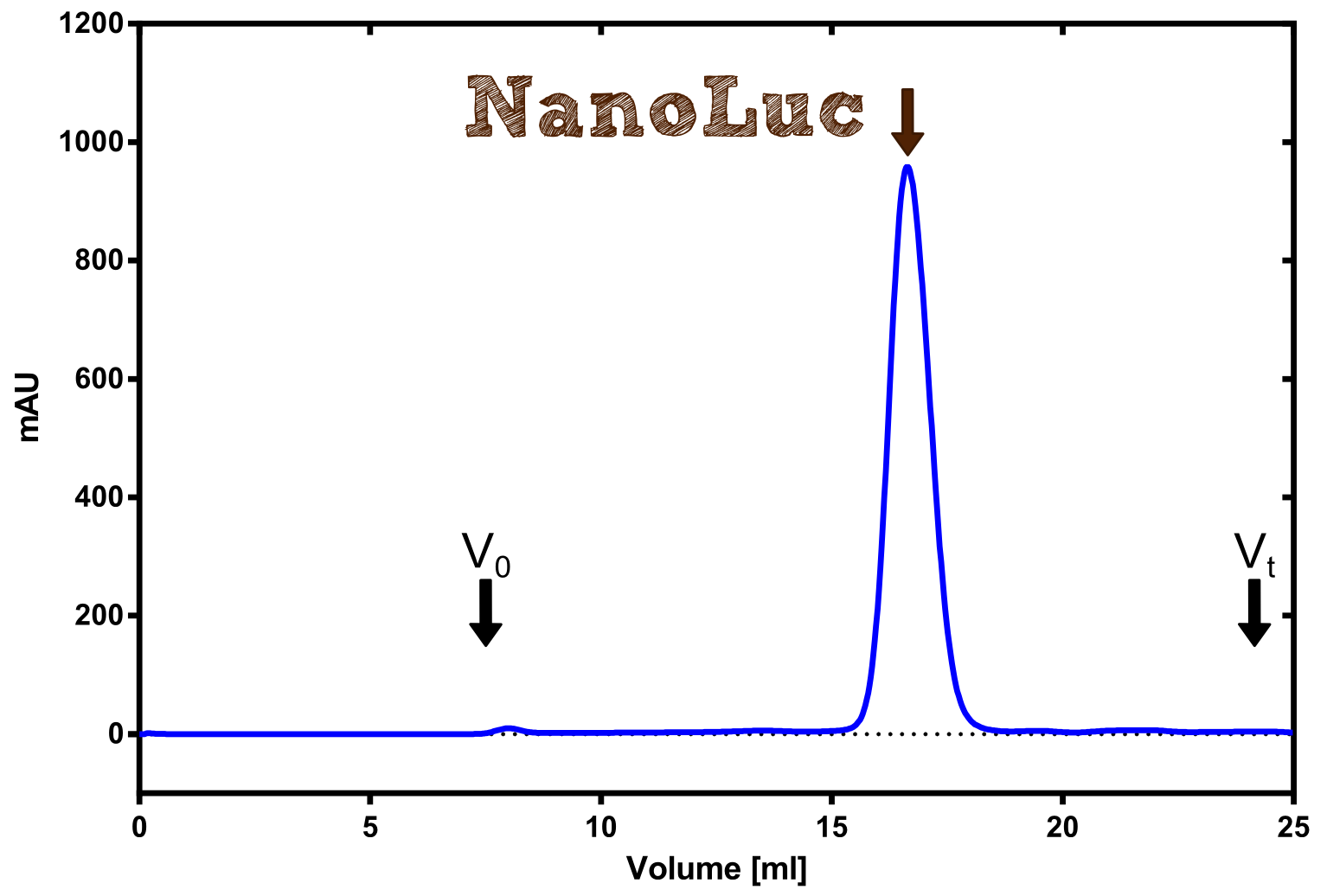
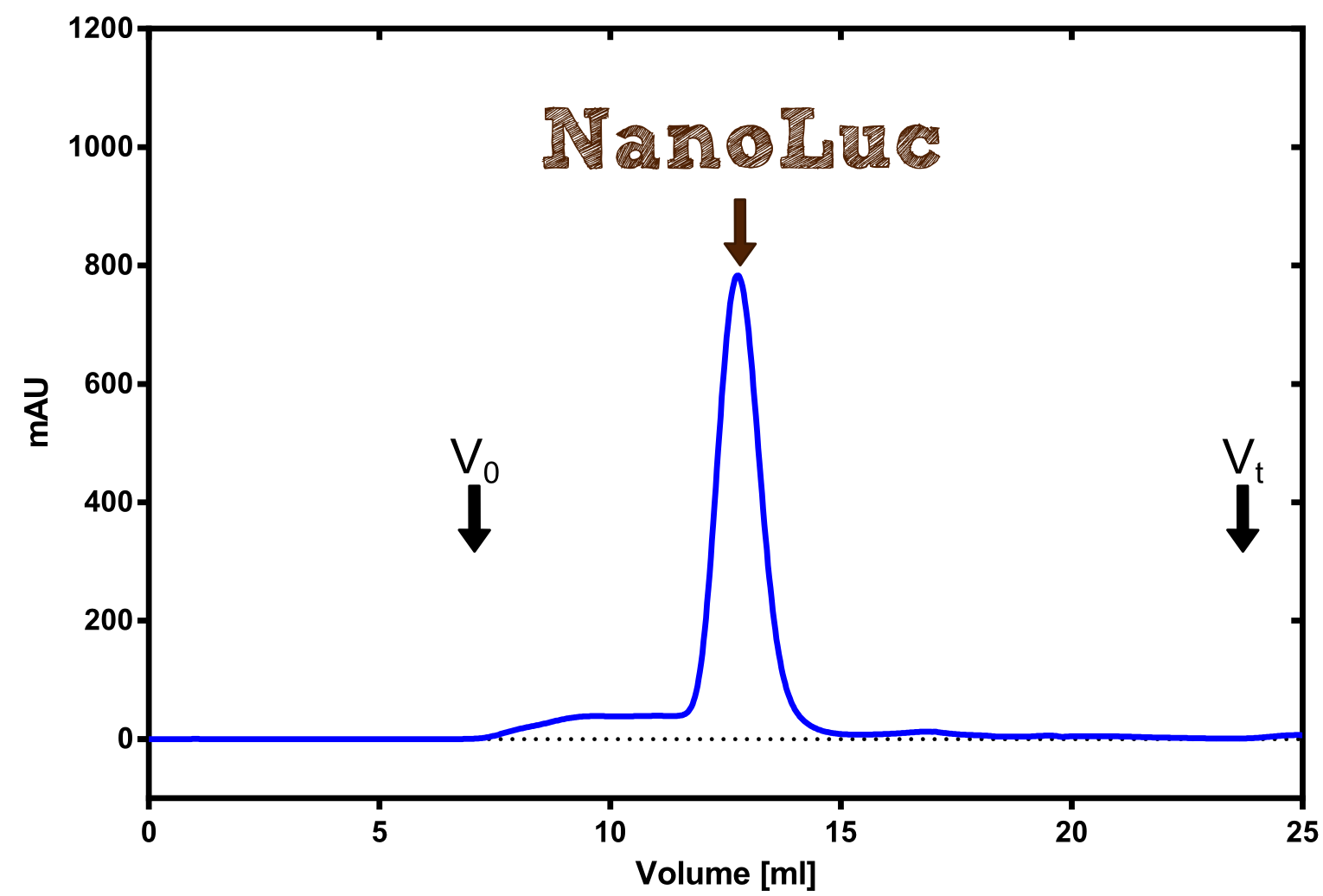
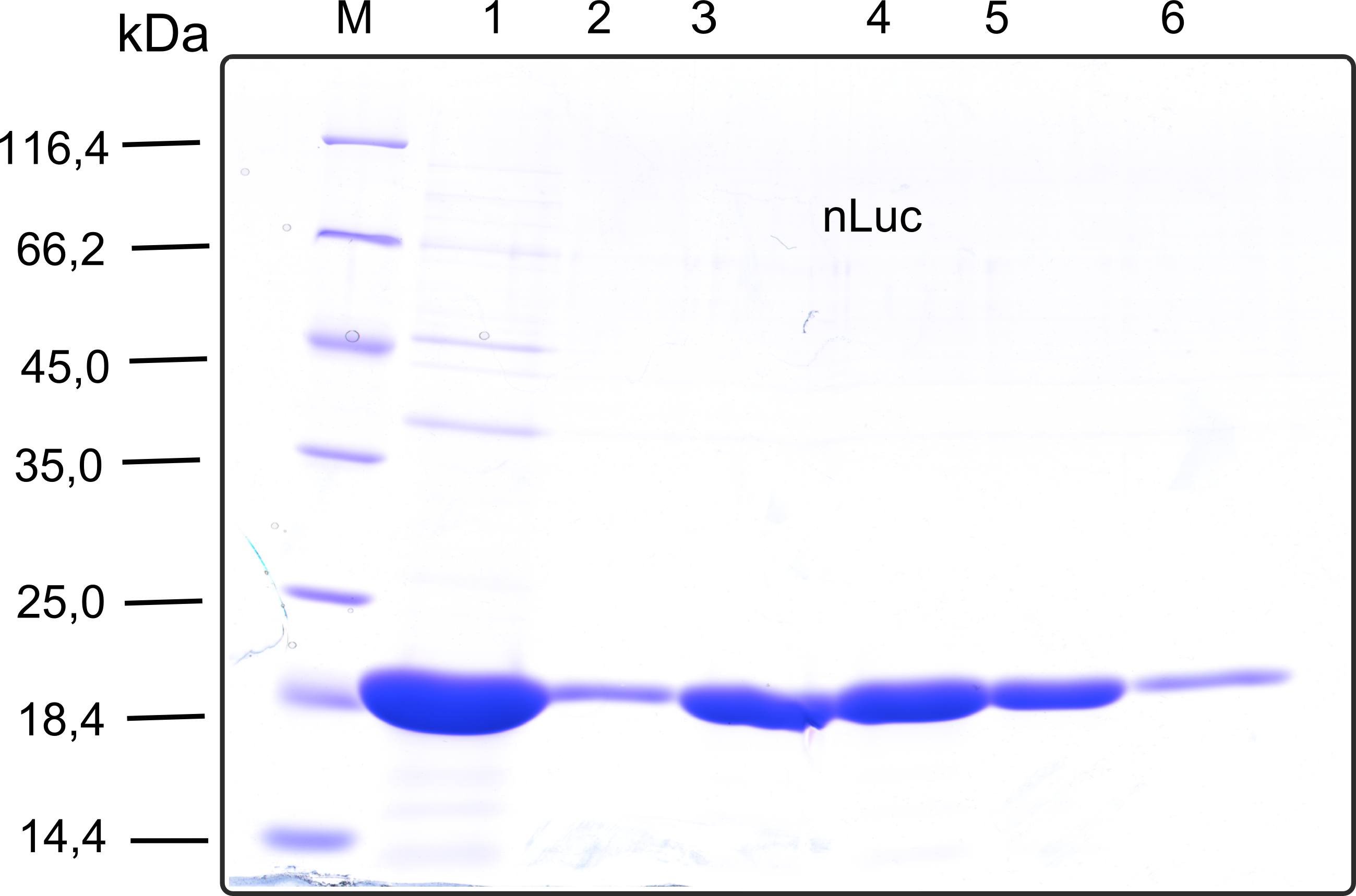
Therefore the NanoLuc was synthesized as a BioBrick in RFC[25] and was produced in E. coli using the pBad expression system with a C-terminal Strep-tag. After the production (2 l of LB-media for analytical and 12 l for preparative preparations) the cells were disrupted using sonification and the lysate was dialysed against 5 l of 1x SA-buffer. Afterwards the lysate was applied to a Streptavidin-Affinity (SA) column and was subsequently washed using SA-Buffer until a baseline was reached and the protein was then eluted using 5 mM of biotin (Attention: These are special columns which are not availible commercially. If you are using commercial colum material you have to use d-Desthiobiotin because usual biotin will elute your protein but you will not be able to regenerate the column after your chromatography). After the SA-chromatography the protein was concentrated using centrifugal concentration units (MWCO: 10 kDa). The concentrated protein was then applied on a Superdex S200/75 size exclusion chromatography. The chromatogram of both preparations show a single peak in the chromatogram which elutes at an expected elution volume of 15 ml. The absence of any notable aggregation peak shows the high stability of this protein and the ease of production.
Structure of the Nano Luciferase
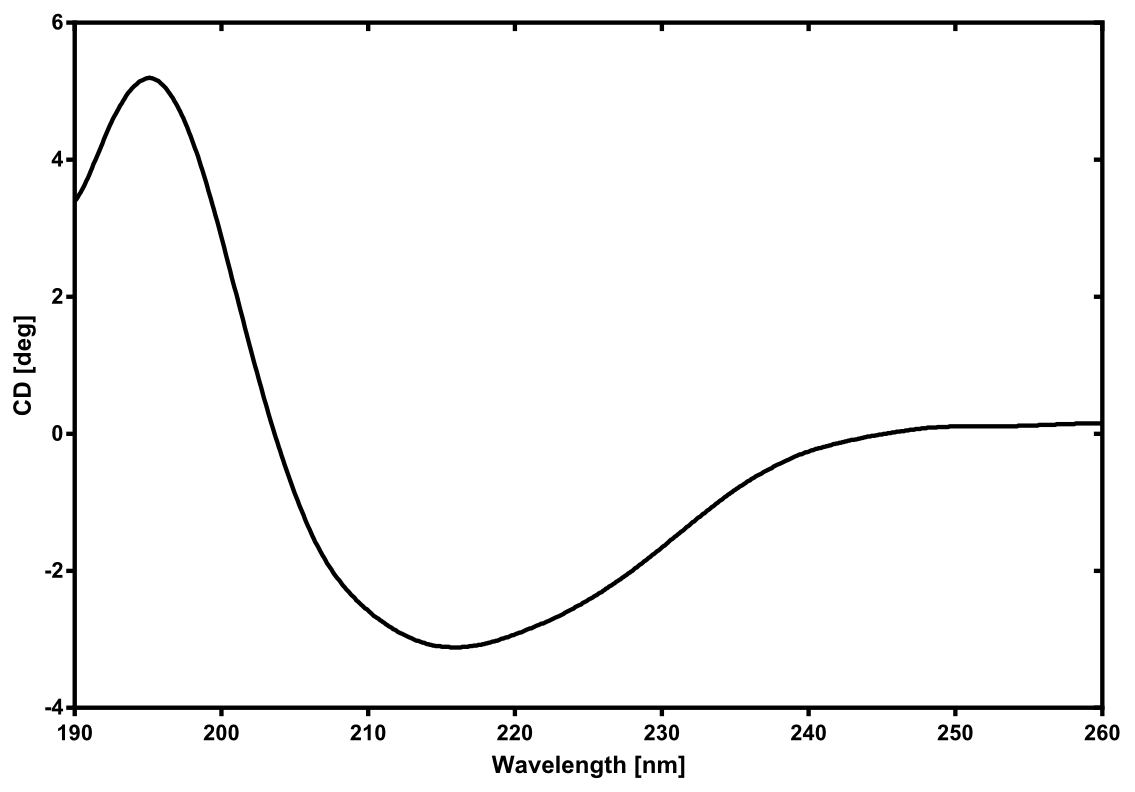
There is no structure availible for the NanoLuc in the [http://www.rcsb.org/pdb/home/home.do Protein Data Bank]. In our [http://2013.igem.org/Team:TU-Munich/Modeling/Protein_Predictions protein modelling] we used homolgy search and identified the structure [http://www.rcsb.org/pdb/explore/explore.do?structureId=3PPT 3ppt_A] as the solved structure with the highest homology to the NanoLuc which has only 21% identify with a similarity of only 0.359. The result of the homology search is shown as annimated gif in Figure 21 (please see our [http://2013.igem.org/Team:TU-Munich/Results/How_To How To] for an introduction). The protein was dialysed against 1x CD-buffer and subsequently a circular dichroism spectroscopy was tanken (learn about [http://2013.igem.org/Team:TU-Munich/Notebook/Methods#Circular_Dichroism_Spectroscopy CD spectroscopy]). The CD spectrum was used to predict the secondary structure content of the NanoLuc which could be determined to 35.1% helix, 27.6% b-strand, 18.5% turn and 18.8% random. As there is only a poor homology present, a detailed comparison of the determined and the predicted secondary structure is not possible. But it can be stated that both show a balanced content of different secondary structures and that the produced protein is present in an folded conformation. The mixed secondary structure content is also in consistance with the predicted secondary structure shown in the [http://2013.igem.org/Team:TU-Munich/Results/Software AutoAnnotator] sequence window (click on show).
Activity determination of Luminescense
The activity of the produced NanoLuc was investigated by its luminescense. The assay was performed in white 96-well plates (Nund) and the bioluminescense was determined at 460 nm in a BioTeK II plate reader for 1 sec per well. For every reaction 50 µl of the NanoGlow substrate (sponsored by Promega) was mixed with 50 µl of the NanoLuc preparation. In a first experiment a dilution series of the NanoLuc was performed in order to find the apropriate dilution of the enzyme (data not shown). Finally the 1:100 000 dilution was used for all experiments as it resulted in intermediate luminescense values within the dynamic range of the plate reader.
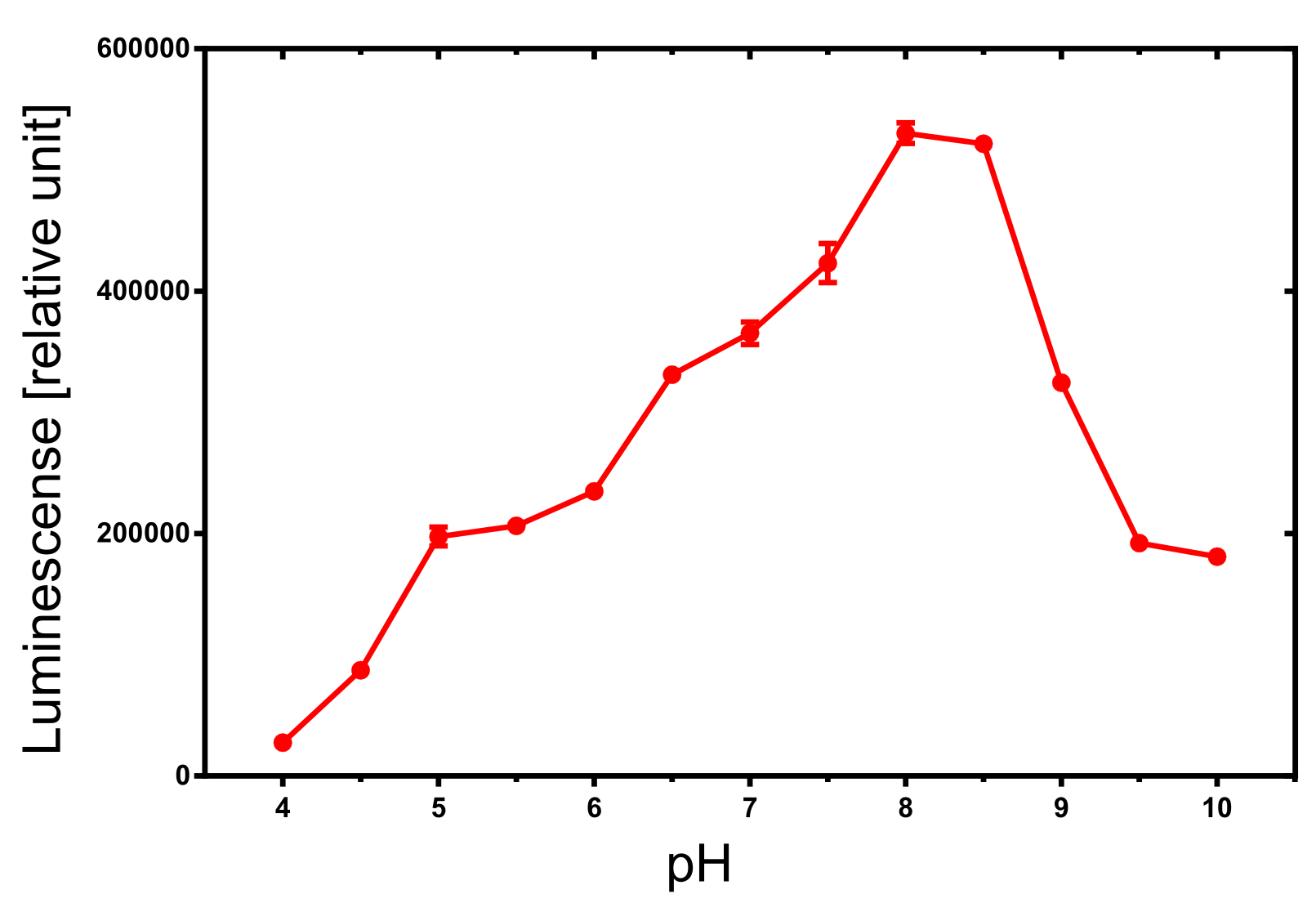
The dependency of the luminescense reaction on the pH value of the buffer was assayed by an experiment in which the NanoGlow substrate was diluted in water instead of the provided buffer and the pH was adapted by 100 µl of 500 mM buffers. The resulting luminescense is shown in Figure x and shows a clear optimum around pH 8.0 to 8.5. A luminescense value of more than 50% can be expected for pH values between 6 and 9 which is sufficient for most assays.
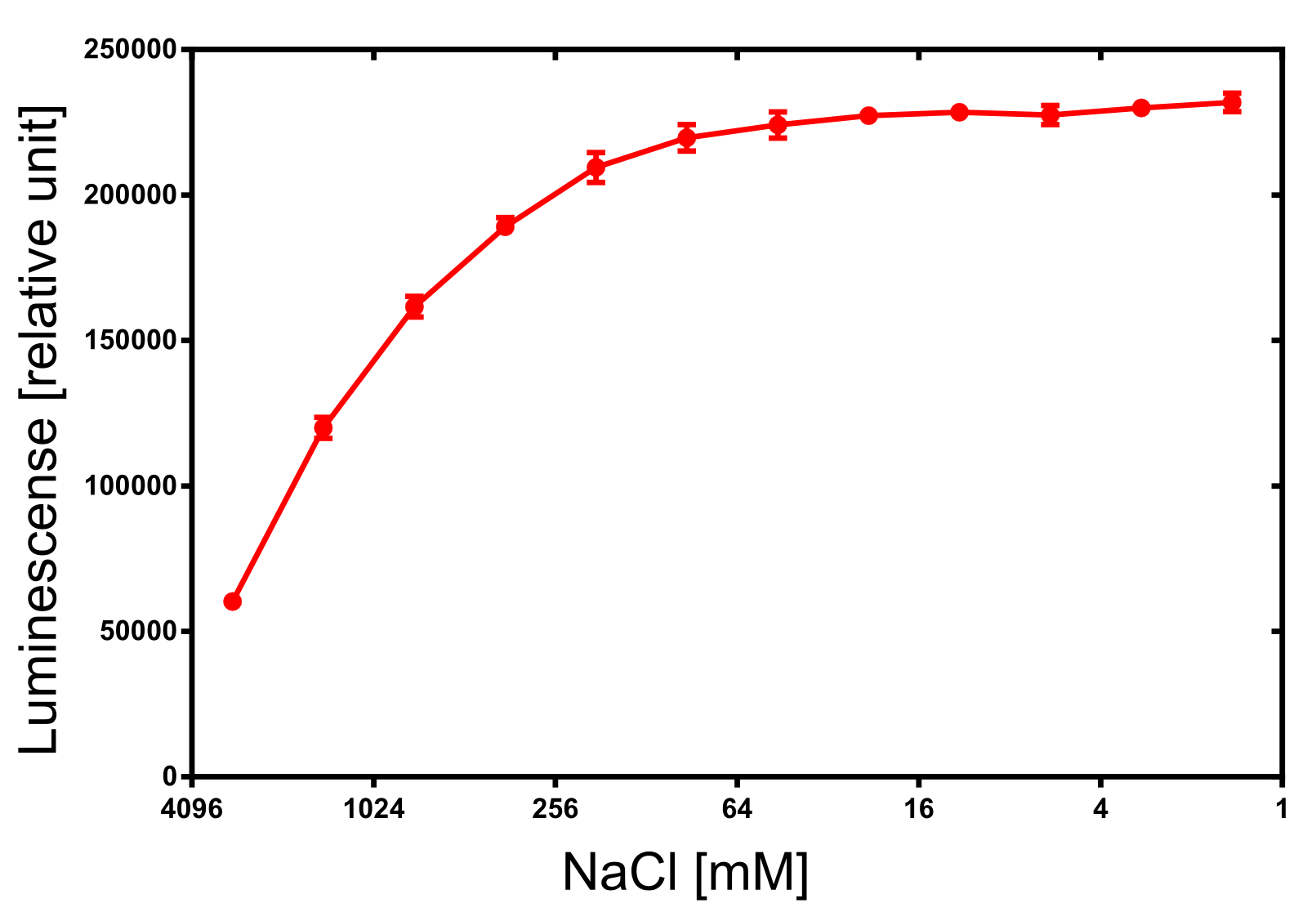
For the dependency of the luminescence on the ionic strength 50 µl of diluted NanoLuc, 50 µl of NanoGlow substrate and 100 µl of sodium chloride dilution series were incubated and afterwards the luminescense was quantified in triplicates. The result is impressing and again speaks for the NanoLuc luciferase as a innovative reporter protein. The obtained luminescense is constant from 0 to 250 mM of NaCl and upon 1 M of NaCl at least 50% of the maximal luminescense can be expected.
Improved by TU_Eindhoven 2019
We developed a cysteine-free version of NanoLuc (BBa_K3168006). By mutating the two cysteines to a serine, the luciferase can be used in fusion proteins that need to be labelled through maleimide coupling, such as our own dCas9-BRET sensor protein (BBa_K3168007).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 10
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]