Difference between revisions of "Part:BBa K1021001:Experience"
(→User Reviews) |
|||
| Line 21: | Line 21: | ||
<!-- DON'T DELETE --><partinfo>BBa_K1021001 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K1021001 EndReviews</partinfo> | ||
| − | + | Team UNIK copenhagen 2015 have also used the nptII resistance gene in experiments. Since the sequence we used is only almost a twin, we had to add it to the registry for the purpose of making a composite biobrick (BBa_K1825005 and BBa_K1825006). Nevertheless, here is our experience using the nptII-gene to transform a new chassis organism. | |
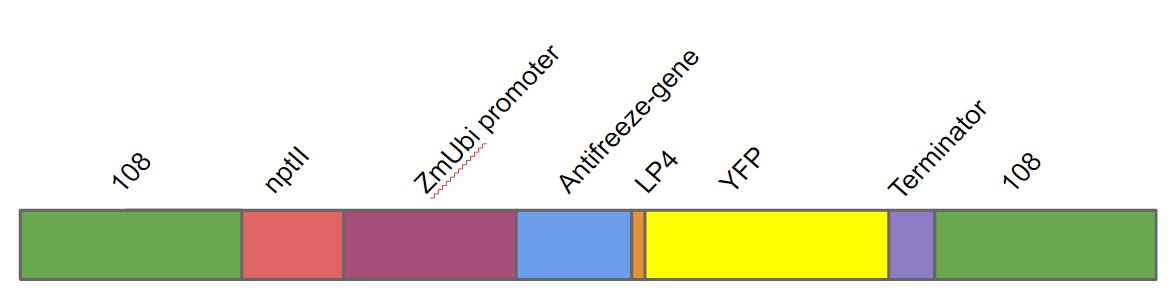
The gene was a part of a larger construct used for transforming ''Physcomitrella patens''. We transformed this gene into ''Physcomitrella patens'' as a part of a larger DNA construct (fig. 1). This DNA construct contained in the following order. A homologous region to the 108 locus on the moss genome (to ensure stable integration into ''P. patens'' genome), the nptII-resistance gene driven by the 35s cauliflower mosaic virus, the Zea mays ubiquitin promoter driving the antifreeze protein (AFP) linked to yellow fluorescent protein (YFP) with the LP4 linker sequence, terminator and lastly another 108 region. | The gene was a part of a larger construct used for transforming ''Physcomitrella patens''. We transformed this gene into ''Physcomitrella patens'' as a part of a larger DNA construct (fig. 1). This DNA construct contained in the following order. A homologous region to the 108 locus on the moss genome (to ensure stable integration into ''P. patens'' genome), the nptII-resistance gene driven by the 35s cauliflower mosaic virus, the Zea mays ubiquitin promoter driving the antifreeze protein (AFP) linked to yellow fluorescent protein (YFP) with the LP4 linker sequence, terminator and lastly another 108 region. | ||
Latest revision as of 15:32, 17 September 2015
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K1021001
- This part is intended to be used as a selective marker for genetic engineering of fungi.
- Geniticin is effective against bacteria as well, so the part can be used in a greater variety of organisms.
User Reviews
UNIQ8002c98988440dc5-partinfo-00000000-QINU UNIQ8002c98988440dc5-partinfo-00000001-QINU
Team UNIK copenhagen 2015 have also used the nptII resistance gene in experiments. Since the sequence we used is only almost a twin, we had to add it to the registry for the purpose of making a composite biobrick (BBa_K1825005 and BBa_K1825006). Nevertheless, here is our experience using the nptII-gene to transform a new chassis organism.
The gene was a part of a larger construct used for transforming Physcomitrella patens. We transformed this gene into Physcomitrella patens as a part of a larger DNA construct (fig. 1). This DNA construct contained in the following order. A homologous region to the 108 locus on the moss genome (to ensure stable integration into P. patens genome), the nptII-resistance gene driven by the 35s cauliflower mosaic virus, the Zea mays ubiquitin promoter driving the antifreeze protein (AFP) linked to yellow fluorescent protein (YFP) with the LP4 linker sequence, terminator and lastly another 108 region.
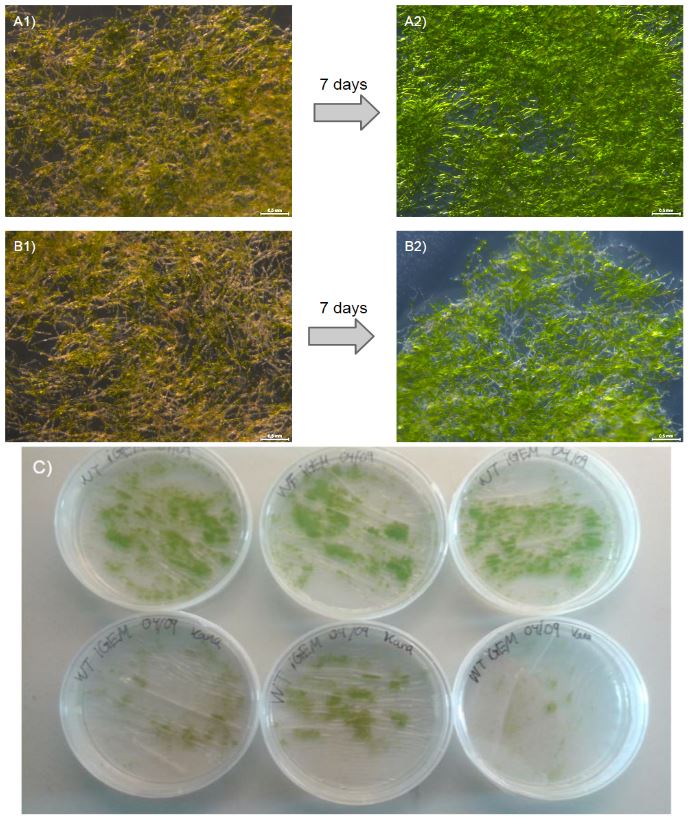
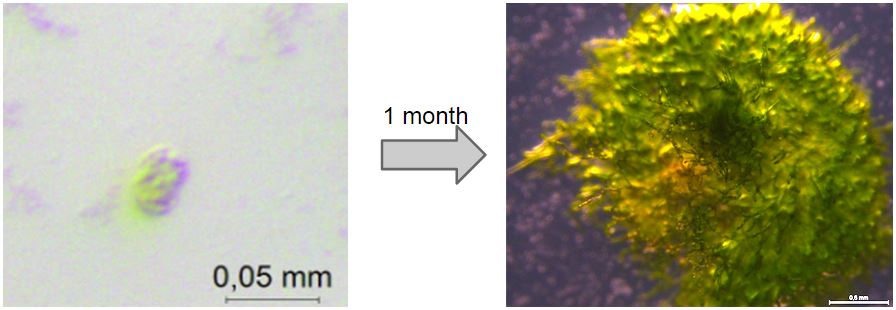
Our moss was able to grow from single transformed protoplasts to full clumps on media containing kanamycin (50 mg/ml), which suggests that the nptII-resistance cassette provides P.Patens with resistance to kanamycin (fig. 2).
To further validate this, we outlined an additional experiment where we grew wild type (WT) moss on either nonselective PhyB-media (3 plates) or PhyB-media containing kanamycin 50 mg/ml (3 plates). After 7 days, there was a visible difference in growth between WT on nonselective media and WT on kanamycin plates (fig. 3). The WT moss planted on kanamycin containing plates grew very little or not at all and is seen under the microscope as withering (fig. 3 B2). This is a stark contrast to our transformed moss that was able to grow from protoplasts to full clumps and the more vibrant WT without kanamycin (fig. 3 A2). This experiment validates the function of the the neomycin phosphotransferase II gene in P. patens.