Chassis/Cell-Free Systems/Commercial E.coli T7 S30/Peak Time
Contents
Commercial E. coli T7 S30
| Cell-Free Systems | Chassis description | Temperature dependence | Peak time | Expression lifespan | Expression capacity |
Peak Time
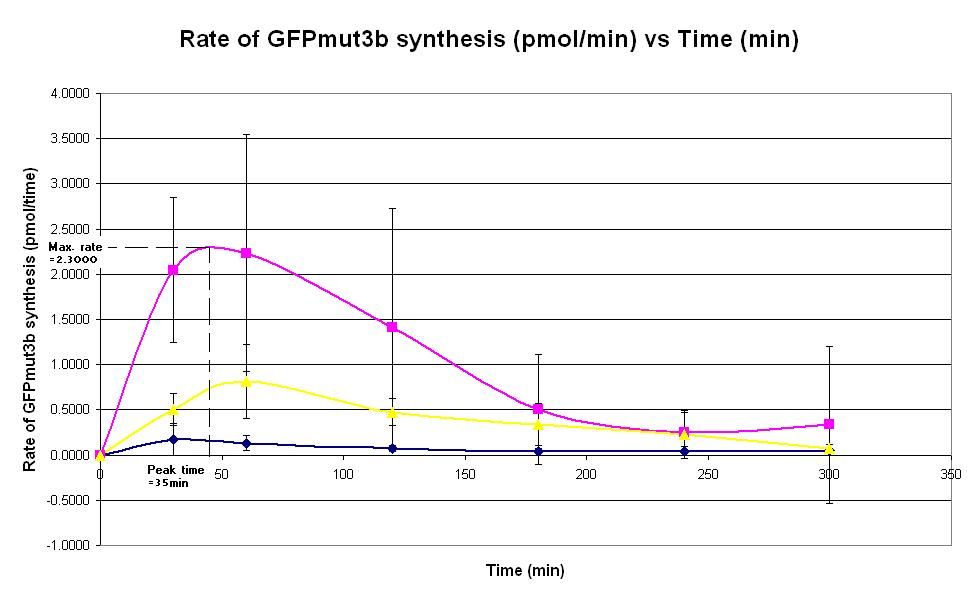
Graph 2. Graph of rate of GFPmut3b synthesis (pmol per min) against time (minutes) at 4oC (dark blue), 25oC (pink) and 37oC (yellow). The simple constitutive gene expression device BBa_I719015 has been used. Each of the three coloured lines show average measurements based on three replicates of the experiment. The error bars represent the standard deviation of the measurements.
Experimental Protocol
Aims
- To determine if the peak time of the commercial E. coli T7 S30 chassis.
- This is done by measuring the fluorescence of the expression system over a 6 hour period, for a range of temperatures from 4oC to 37oC.
Equipment
- Fluorometer + Connected PC
- 3 Fluorometer Plates (Black)
- Sticky Seal Tape
- Eppendorf Tubes
- Gilson p20,p200,p1000
- Stop watch
- Foil
Reagents
- Commercial S30 E.coli extract. Including:
- 175µl Amino Acid Mixture Minus Cysteine, 1mM
- 175µl Amino Acid Mixture Minus Methionine, 1mM
- 175µl Amino Acid Mixture Minus Leucine, 1mM
- 450µl S30 Extract, Circular (3 × 150µl)
- 750µl S30 Premix Without Amino Acids
- Nuclease Free water x1ml
- DNA pT7-GFP from midiprep
- DNA pT7 from midiprep
Preparation
- First collect all equipment and reagents and ensure that the fluorometer and that the PC connected has a data collection protocol installed.
- Turn on the water bath at 25 °C and 37 °C incubator.
- Commercial E.coli Cell Extract: First prepare cell extract for 13 reaction. Add the 32.5μl volume of two amino acid minus mixtures into an labeled eppendorf to give a volume of 66μl. Each amino acid minus mixture is missing one type of amino acid, and so by combining two solutions we are complementing each solution for the missing amino acid. Place eppendorf in a rack on bench. Take an eppendorf tube and add 260µl of S30 Premix. Then add 195µl of S30 Extract Circular.
- DNA constructs We need to prepare a dilution of DNA to give 4ug of DNA. Add 64 of pT7-GFP DNA to 136ul of Nuclease Free water (10 reactions worth). Add 39.2 ul of pT7 DNA to 20.8ul of Nuclease Free Water (4 reactions worth)
Plate loading
- To each of the plate, place the cell extracts in 4 wells, and add the pT7-GFP to 3 of the wells, and pT7 DNA to the last well. Then top up each well to a total volume of 60ul.
- Place the plate in the fluorometer to measure its initial fluorescent reading.
- After the measurement, place the sticky tape across the plate, and put the plate in the 37oC water bath.
- Start on the next plate, and place the plate in the 25oC water bath.
- Repeat the same procedures for the remaining plate at 4oC.
- Before placing them in the water bath, wrap aluminium foil around the plates to prevent photobleaching.
- Stagger the start of all the plates by around 5 minutes.
- Repeat the measurements every hour, for 6 hours.
- Using the [http://2007.igem.org/Imperial/Wet_Lab/Results/Res1.3 GFPmut3b calibration curve], plot a graph of rate of GFPmut3b synthesis against time for all 3 plates.
Results
Our raw data and data analysis can be found here: File:T7 Data.xls
