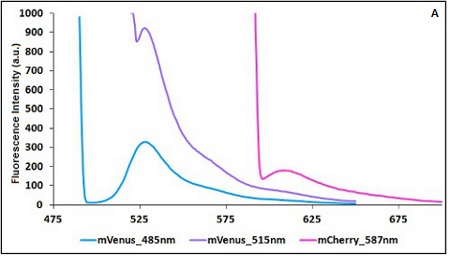
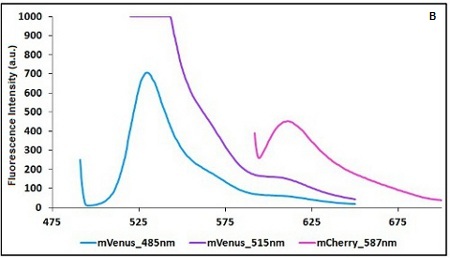
BBa K731700 and BBa K731710 measurements
Hello world
In this page we are really proud to introduce you the protocol we (Giacomo and Anna) developed for the characterization transcriptional terminators effects on gene expression. (Tested on BBa_J64997)
The develop of this protocolcost us not only sleepless night but also sunny and beautiful weekend of trekking so take a hot cup of tea and read it all.
IN VIVO ANALYSIS
|
BEFORE STARTING:
NOTE1:Antibiotic are at concentration of 0.1mg/ml ampicillin and 0.035mg/ml cloranphenicol NOTE2:Four glycerol stocks for both control and sample will give you a good level of significance
|
PROTOCOL DEVELOPMENT:
Emission and Excitation parameter choise:
The emission and excitation parameters must be aimed at achievement of the maximum distance between the two points at which you take emission measurements and the maximum comparability in terms of fluorescence intensity according to the limits imposed by the two fluorescent proteins in use.
Example: The two proteins in use in BBa_K731700 and BBa_K731710 are mCherry and A206K Venus (mVenus). After excitation of the two proteins with standard parameters we note a huge difference in fluorescence intensity between the two proteins; moreover the proximity between the excitation wavelenght and the emission peak can easily influence the measurements.
In this case a shift of the brighter protein (mVenus) excitation wavelenght toward smaller wavelenght can bring several benefits:
- A decrease in fluorescene intensity
- A larger distance between the excitation wavelenght and the emission peak
The same approach for the less bright protein (mCherry) is not exploitable since would have the effect of further reducing the emission peak intensity; the better way to have a lower influence on the emission peak due to the proximity to the excitation wavelenght is to use a wavelenght different from the emission peak for measurements (point with a bigger wasvelenght).
| Standard Excitation (nm) | Standard Emission (nm) | Modified Excitation (nm) | Modified Emission (nm) | |
| mCherry | 587 | 609 | 587 | 615 |
| mVenus | 515 | 528 | 485 | 528 |
TABLE 1. Standard and modified excitation and emission wavelengths
CHART 1A/B: Emission scan of BBa_K731700 (A) and BBa_K731710 (B) with standard and modified excitation wavelenght (instrument: Varian Cary Eclipse, voltage: 570 V (A) and 520 V (B)
Sonication of the samples:
In order to reduce the scattering (signal due to excitation light in proximity of the excitation wavelenght) and to have a clear signal, we decided to sonicate our sample before measure them. Both the improvement are due to the lower O.D. of a sonicated sample if compared to the one of a cell suspension sample. A bigger O.D. means in fact a decrease in the amount of light emitted by the fluorescent proteins that reach the sensor (light that), a decrease in the light that actually reach the fluorescent proteins and an icrease of the excitation light that is reflected to the sensor (increase in scattering).