Difference between revisions of "BBa K731700 and BBa K731710 measurements"
| Line 111: | Line 111: | ||
<p style="width:900px; margin-bottom:60px; text-align:justify"><em> | <p style="width:900px; margin-bottom:60px; text-align:justify"><em> | ||
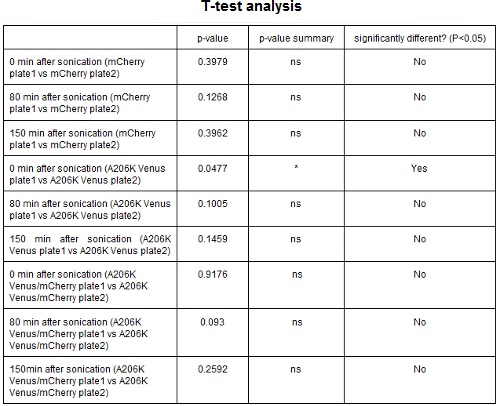
| − | <strong>CHART 3:</strong> '' | + | <strong>CHART 3:</strong> ''Fluorescence emission raw peaks of two cultures from colonies from different plates.''<br/> |
| − | <strong>TABLE 2:</strong> ''The table shows the statistic analysis performed on the data in CHART 3. Neither the proteins peaks of the two plates nor their ratio are significantly different (the result of | + | <strong>TABLE 2:</strong> ''The table shows the statistic analysis performed on the data in CHART 3. Neither the proteins peaks of the two plates nor their ratio are significantly different (the result of "0 min after sonication (A206K Venus plate 1 vs A206K Venus plate 2)" is considered a false positive since the other two times show no significant difference).''<br/> |
</em> </p> | </em> </p> | ||
'''Different concentrations of inducing agent:''' | '''Different concentrations of inducing agent:''' | ||
| − | The use of different concentration of inducing agent should not influence the ratio between the fluorescence intensity of the two proteins. Nevertheless | + | The use of different concentration of inducing agent should not influence the ratio between the fluorescence intensity of the two proteins. Nevertheless the inducing agent concentration may have influence on the quantity of proteins synthesized. We tested two different concentration of isopropyl β-D-1-thiogalactopyranoside (IPTG). |
| − | |||
[[Image:IPTGconcchart_inductiontime.jpg]][[Image:Statiptg.jpg]] | [[Image:IPTGconcchart_inductiontime.jpg]][[Image:Statiptg.jpg]] | ||
| Line 126: | Line 125: | ||
<p style="width:900px; margin-bottom:60px; text-align:justify"><em> | <p style="width:900px; margin-bottom:60px; text-align:justify"><em> | ||
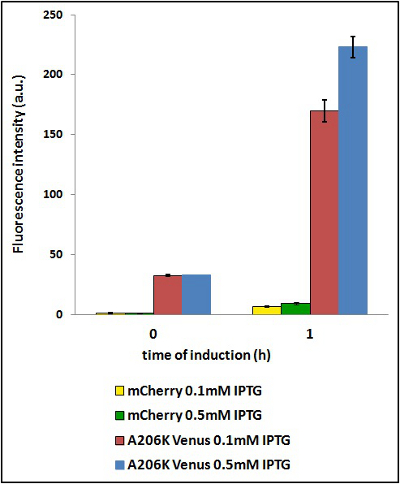
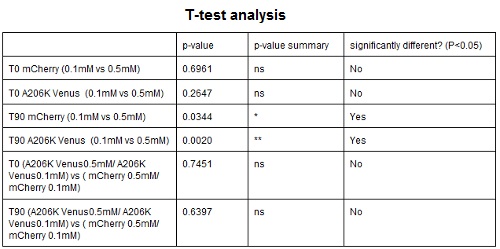
| − | <strong>CHART 4:</strong> '' | + | <strong>CHART 4:</strong> ''Raw emission peaks of coltures iduced with 0.1 mM or 0.5 mM IPTG. The measurements were taken just after induction (time 0) and after 90 minutes (time 1).''<br/> |
| − | <strong>TABLE 3:</strong> ''The table shows the statistic analysis permormed on the data in CHART 4. There is no significant | + | <strong>TABLE 3:</strong> ''The table shows the statistic analysis permormed on the data in CHART 4. There is no significant difference between the two samples with different IPTG concentration at time 0; significant differences are instead shown between the samples at induction time 90 min. The t-tests also show that the % increase in protein expression between 0.1mM e 0.5mM is the same in the two proteins. ''<br/> |
</em> </p> | </em> </p> | ||
Revision as of 08:02, 25 September 2012
Hello world
In this page we are really proud to introduce you the protocol we (Giacomo and Anna) developed for the characterization transcriptional terminators' effects on gene expression. (Tested on BBa_J64997)
The develop of this protocol cost us not only sleepless night but also wasted sunny and beautiful weekend so take a hot cup of tea and read it all.
IN VIVO ANALYSIS
|
BEFORE STARTING:
NOTE1:Antibiotic are at concentration of 0.1mg/ml ampicillin and 0.035mg/ml cloranphenicol NOTE2:Four glycerol stocks for both control and sample will give you a good level of significance
|
PROTOCOL DEVELOPMENT:
Emission and Excitation parameter choise:
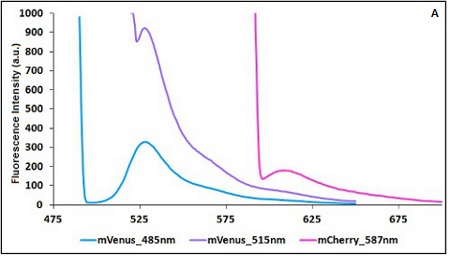
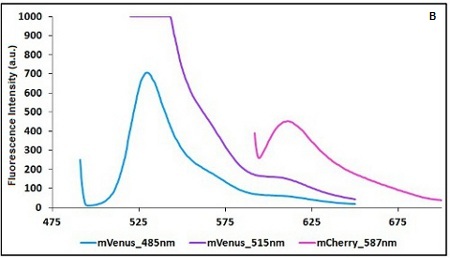
The two proteins in use in BBa_K731700 and BBa_K731710 are mCherry and A206K Venus (mVenus), respectively upstream and downstream of the terminator. After excitation of the two proteins with standard parameters, we noticed a huge difference in fluorescence intensity between the two peaks, as mVenus emission peak was about 10 times stronger than the mCherry peak. Moreover, the two fluorescent proteins presented excitation and emission peaks very close to each other. This feature may cause unwanted interferences of the scattering effect caused by the excitation ray. // Due to the scattering effect caused by the closeness of the two peaks, interferences in the measure can occur.
In this case a shift of the brighter protein (mVenus) excitation wavelength toward smaller wavelength can bring two main benefits:
- A decrease in emission peak's fluorescence intensity
- A larger gap between the excitation wavelength and the emission peak's wavelength
Moreover, the less bright protein (mCherry) was measured at a slightly higher wavelength than the peak seen in the emission spectrum.
Here is a summary of the wavelengths used to take the measurements with BBa_K731700 and BBa_K731710:
| Standard Excitation (nm) | Standard Emission (nm) | Modified Excitation (nm) | Modified Emission (nm) | |
| mCherry | 587 | 609 | 587 | 615 |
| mVenus | 515 | 528 | 485 | 528 |
TABLE 1: Standard and modified excitation and emission wavelengths
CHART 1A/B: Emission scan of BBa_K731700 (A) and BBa_K731710 (B) with standard and modified excitation wavelenght. The measurements were taken with a Varian Cary Eclipse spectrofluorimeter, with 5nm slit and a voltage of 570 V (A) and 520 V (B))
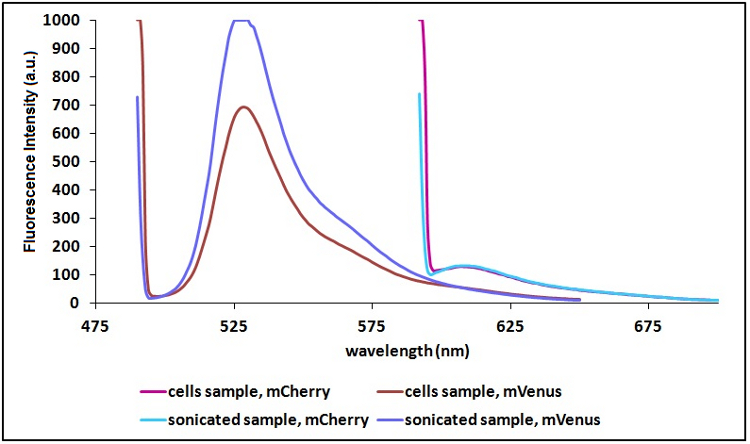
Sonication of the samples:
In order to reduce the scattering due to excitation ray and thus to have a clearer signal, we decided to sonicate our sample before measure them. Both the improvements are due to the lower O.D. of a sonicated sample compared to a cell suspension sample. A higher O.D. causes a decrease in light intensity emitted by the fluorescent proteins that reach the sensor, a decrease in the exciting light that actually reach the fluorescent proteins and an increase in the excitation light that is reflected to the sensor (increase in scattering).
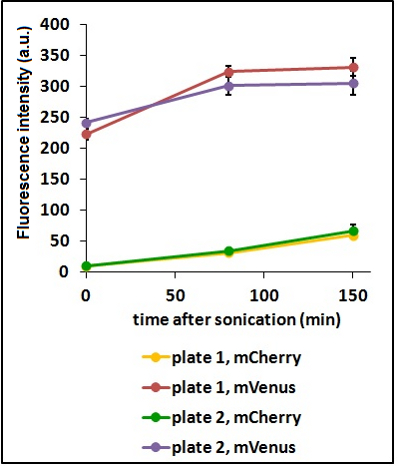
Test on different colonies:
The use of glycerol stock instead of O.N cultures makes the procedure easier and faster. However, glycerol stocks come from the same colony; to verify the consistency of our results we tested some different colonies from two different plates.
CHART 3: Fluorescence emission raw peaks of two cultures from colonies from different plates.
TABLE 2: The table shows the statistic analysis performed on the data in CHART 3. Neither the proteins peaks of the two plates nor their ratio are significantly different (the result of "0 min after sonication (A206K Venus plate 1 vs A206K Venus plate 2)" is considered a false positive since the other two times show no significant difference).
Different concentrations of inducing agent:
The use of different concentration of inducing agent should not influence the ratio between the fluorescence intensity of the two proteins. Nevertheless the inducing agent concentration may have influence on the quantity of proteins synthesized. We tested two different concentration of isopropyl β-D-1-thiogalactopyranoside (IPTG).
CHART 4: Raw emission peaks of coltures iduced with 0.1 mM or 0.5 mM IPTG. The measurements were taken just after induction (time 0) and after 90 minutes (time 1).
TABLE 3: The table shows the statistic analysis permormed on the data in CHART 4. There is no significant difference between the two samples with different IPTG concentration at time 0; significant differences are instead shown between the samples at induction time 90 min. The t-tests also show that the % increase in protein expression between 0.1mM e 0.5mM is the same in the two proteins.
O.D. equalization:
This point allows to have low standard deviation and variability in the data.