Part:BBa_C0062
luxR repressor/activator, (no LVA?)
In complex with HSL, LuxR binds to the Lux promoter, activating transcription from Pr BBa_R0062, and repressing transcription from Pl BBa_R0063.
The lux cassette of V. fischeri contains a left and a right promoter. The right promoter gives weak constitutive expression of downstream genes.This expression is up-regulated by the action of the Lux activator, LuxR complexed to HSL. Two molecules of LuxR protein form a complex with two molecules the signalling compound homoserine lactone (HSL). This complex binds to a palindromic site on the promoter, increasing the rate of transcription.
Usage and Biology
We improved this part by using it in a part which allows for the constitutive expression of LuxR (BBa_K1893004). This part also includes the pLux promoter, so that genes placed downstream the promoter will be expressed when induced with 3OC6-AHL. We also made a part which allows for the characterisation of LuxR, as we placed GFP downstream the pLux promoter (BBa_K1893005). The part is currently being characterised.
Unspecified.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
- Allergen characterization of BBa_C0062: Not a potential allergen
The Baltimore Biocrew 2017 team discovered that proteins generated through biobrick parts can be evaluated for allergenicity. This information is important to the people using these parts in the lab, as well as when considering using the protein for mass production, or using in the environment. The allergenicity test permits a comparison between the sequences of the biobrick parts and the identified allergen proteins enlisted in a data base.The higher the similarity between the biobricks and the proteins, the more likely the biobrick is allergenic cross-reactive. In the full-length alignments by FASTA, 30% or more amount of similarity signifies that the biobrick has a Precaution Status meaning there is a potential risk with using the part. A 50% or more amount of identity signifies that the biobrick has a Possible Allergen Status. In the sliding window of 80 amino acid segments, greater than 35% signifies similarity to allergens. The percentage of similarity implies the potential of harm biobricks’ potential negative impact to exposed populations. For more information on how to assess your own biobrick part please see the “Allergenicity Testing Protocol” in the following page http://2017.igem.org/Team:Baltimore_Bio-Crew/Experiments
For the biobrick Part BBa_C0062, there was a 0% of identity match and 0% similarity match to the top allergens in the allergen database. This means that the biobrick part is not of potential allergen status. In 80 amino acid alignments by FASTA window, no matches found that are greater than 35% for this biobrick. This also means that there is not of potential allergen status.
>Internal Priming Screening Characterization of BBa_C0062: Has 1 possible internal priming site between this BioBrick part and the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
For the BioBrick part BBa_C0062, the first location of the internal priming site is on the 628-634 base number of the BioBrick and on the 14-20 base number of the VR primer.
Contribution: SiCAU-China 2017
Authors:Wenwen Lin
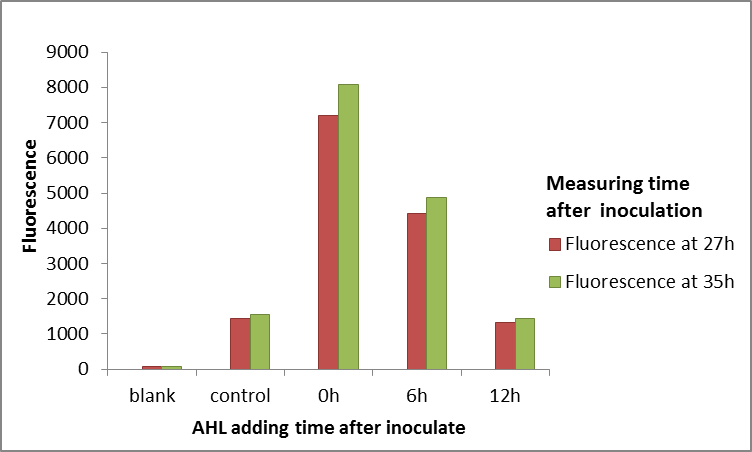
Summary:The QS system LuxR/LuxI has been widely used in kinds of logic circuit. But this year we find that LuxR can not always work well in E.coli. The LuxR, in E.coli, will not work when E.coli enter into stationary phase. This characteristic may be useful to users who will use LuxR as an activating transcription. (more details)
Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary:
We adapted the part to be able to assemble transcriptional units with the Golden Gate assembly method
Documentation:
In order to create our complete [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Printeira Part Collection] of parts compatible with the Golden Gate assembly method, we have made the part BBa_K2656016 which is BBa_C0062 adapted to the Golden Gate Technology.
HZAU-China 2019's Characterization
Design
In order to fit the project and guide the experiment, our research object for characterization studies is LuxR protein.
The plasmid consists of promoter (one of J23102, J23105, J23109), RBS B0032, luxR, terminator B1002, luxPR promoter, gfp coding sequence E0040,rrnB T1 terminator and T7Te terminator which are arranged in an order on a pSB1C3 backbone. We aim to induce the transcription of the downstream GFP by luxPR promoter by artificially adding different concentrations of HSL and LuxR which can form an activator complex LuxR-HSL. The amount of LuxR-HSL can be detected by luxPR promoter. The higher intensity of GFP expressed by luxPR, the more LuxR-HSL complex there will be. In this way, we can get insight into the binding characteristics of LuxR and HSL.
Within 15 hours after the addition of HSL, the OD600 and fluorescence intensity were measured using a microplate reader.
Expectation
Fluorescence is expected to occur in all three cultures, as all of these include the GFP coding sequence. However, due to the different intensities of the three promoters , J23102, J23105 and J23109, we predict that the fluorescence intensity of the same promoter should increase with the increase of HSL concentration, and the fluorescence intensity at the same HSL concentration should increase as the intensities of the promoter arise.
Materials and Methods
Plasmids Construction
luxR, luxPR and gfp are directly taken from the iGEM 2019 distribution kit and the promoter, RBS and terminator are ligated between the parts by overlap extension PCR, and the constructed fragment is ligated to PSB1C3 by homologous recombination. The correct construction is confirmed by sequencing.
Inoculation and cultivation
1.Pick three isolated colonies with a sterile tip from each of the three LB plates and add each tip into 5mL LB medium with 170μg/mL chloramphenicol. Incubate overnight at 37℃ in a shaker with 200rpm. Take 100μL bacterial of each flask into 5mL fresh new LB and incubate for another 3-4 hours.
2.Add each kind of bacterial fluid into different lines of a sterile, black-coated 96-well plate. Add shaken bacterial solution and fresh medium with 170μg/ml chloramphenicol to a total of 100μL, and make sure the OD600 of each well is 0.05-0.08, measured by the plate reader.
3.Add 1μL HSL diluted in DMSO into each well. The final concentration range of HSL is from 10-8 to 0.1 mmol/L. Add 1μL DMSO without HSL served as positive controls. Blank controls are fresh LB medium with DH5α. Place the 96-well plate into the automatic microplate reader (Synergy H1 hybrid multi-mode reader). Incubate at 37℃ overnight and measure the fluorescence value(excitation: 485nm, emission: 528nm)and OD600 of each well every 30 minutes.
4.Data taken from the plate reader are exported to Excel and imported to Origin for analysis.
5.All experiments above are carried out in 3 biological replications.
Results and analysis
Due to the different expression levels of GFP, the metabolic pressure of bacteria is different, and the growth of bacteria with different constitutive promoters is different. Therefore, it is difficult to find a time point of the bacteria to achieve proper data between different constructs. Finally, we take the 11th hour after adding HSL to collect data since the bacteria in most groups are in platform stage or just have slight decrease.

Figure 1. Fluorescence/OD600 over the concentration of HSL. Data are collected after 11 hours’ induction. All experiments are carried out in three biological replications and the error bar indicates standard error.
From figure 1, for each constitutive promoter, the ratio of fluorescence intensity to OD600 increases as the concentration of HSL increases after 11 hours’ induction (Figure 1). As known to everyone, J23102 has the highest transcription intensity followed by J23105, and J23109 remains the lowest. When the concentration of HSL is higher than 101nM, the ratio of fluorescence intensity to OD600 increases substantially as the intensities of the promoters increase at the same HSL concentration. At higher concentrations of HSL, the ratio of fluorescence intensity to OD600 decreases abnormally, most likely due to excessive metabolic stress induced by high concentration of HSL. From figure 2 we can figure out that high concentration of HSL has put excessive metabolism pressure on the bacteria. Because, it is 103-104nm/L HSL that induce the highest fluorescence intensity. As the concentration of HSL reaches 105nm/L, the intensity significantly decreases, even becomes lower than the intensity of 102nm/L (Figure 2).

Figure 2. Dynamic curves of fluorescence over time. A. Fluorescence intensity over time at different concentrations of HSL of J23102. B. Fluorescence intensity over time at different concentrations of HSL of J23105. C. Fluorescence intensity over time at different concentrations of HSL of J23109. All experiments are carried out in three biological replications and the error bar indicates standard error.
In conclusion, the binding ability of LuxR and HSL is strong, and only a few LuxR are needed to respond to HSL of different concentrations. Moreover, a high concentration of HSL will lead to excessive metabolic pressure of bacteria, so an appropriate concentration of HSL is very necessary. The most appropriate concentration of HSL is about 103 nM as we determined.
Characterization by 2019 iBowu-China
We character the ability of this part(LuxR) binding to N-(3-oxohexanoyl)-L-homoserine lactone (OHHL,quorum sensing signal molecular of E.carotovora) in vivo and vitro system.
Results
1.The report system we used
We used promoter J23101 to promoter LuxR, and its binding promoter pLux(BBa_R0062) driving GFP (BBa_I746916) as a reporter to character its binding ability to OHHL(fig1).

Figure 1. gene circuits of reporter
2.Results in vivo

Figure 2.The fluorescence intensity over the time with different concentration of OHHL treatment. The inducer OHHL was added to the culture after 60 min incubation.
From figure2, we found that the GFP intensity increased after OHHL incubation, even 1nM OHHL, which means LuxR could bind to OHHL efficiently. It means that LuxR-pLux could be a highly sensitive detector of OHHL.
3.Results in vitro(cell-free system).
After the cellular sensing characterization, we test the binding efficiency of LuxR to OHHL in cell-free systems(Fig. 3). After the overnight cell-free incubation, it showed that the circuit of LuxR-GFP was responsible to AHL with the detection limit of 1 nM.

Figure 3. The cell-free fluorescence output induced by a series of AHL concentrations. FLU was measured after overnight incubation
ShanghaiTech_China 2022's Characterization
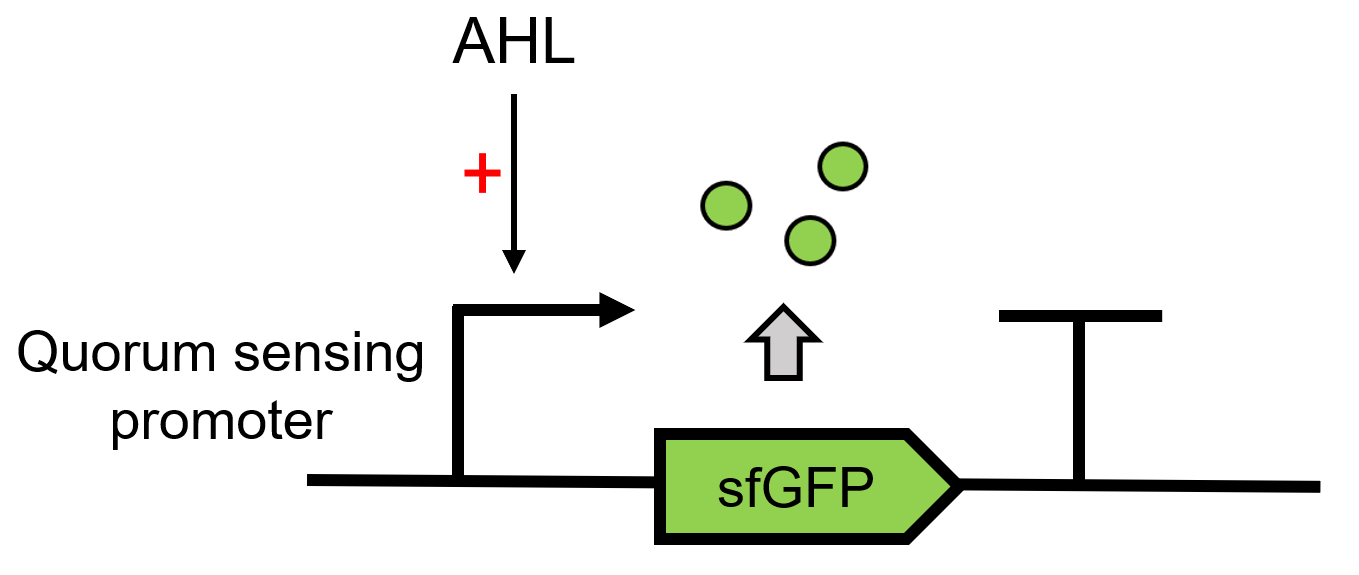
Design
LuxR receives the signal of AHL, which is sent by LuxI(BBa_C0161), 3OC6 HSL here. It can then regulate the downstream Lux pR (BBa_R0062) to make superfolder GFP (BBa_K4115000) express.(Figure 1)
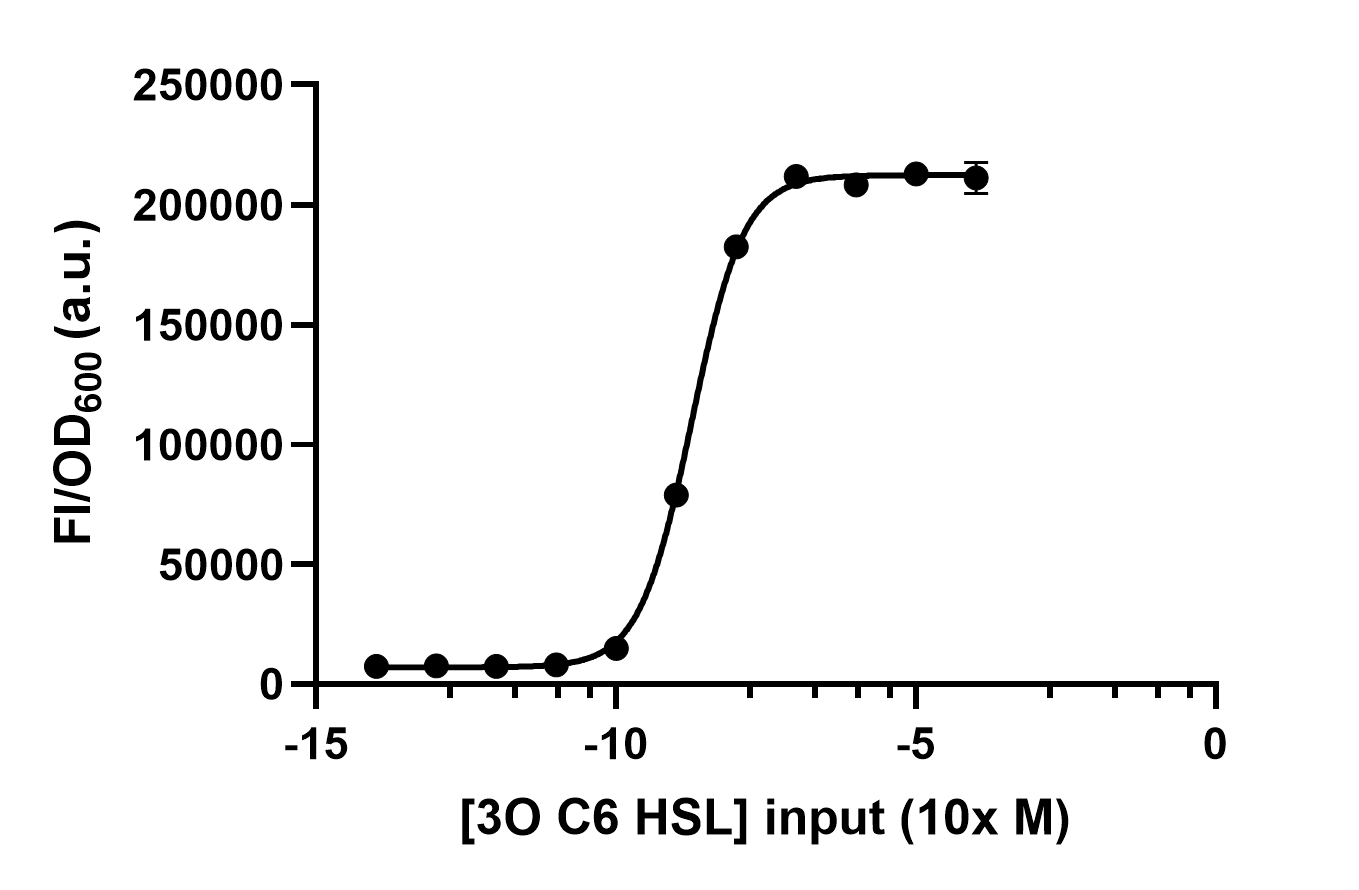
Results
We use different concentrations of the autoinducer, ranging from 10-4 to 10-14 M, to test the response intensity of LuxR.The result is shown by dividing the fluorescence intensity by the OD600, both measured in the transformed E. coil. (Figure 2).
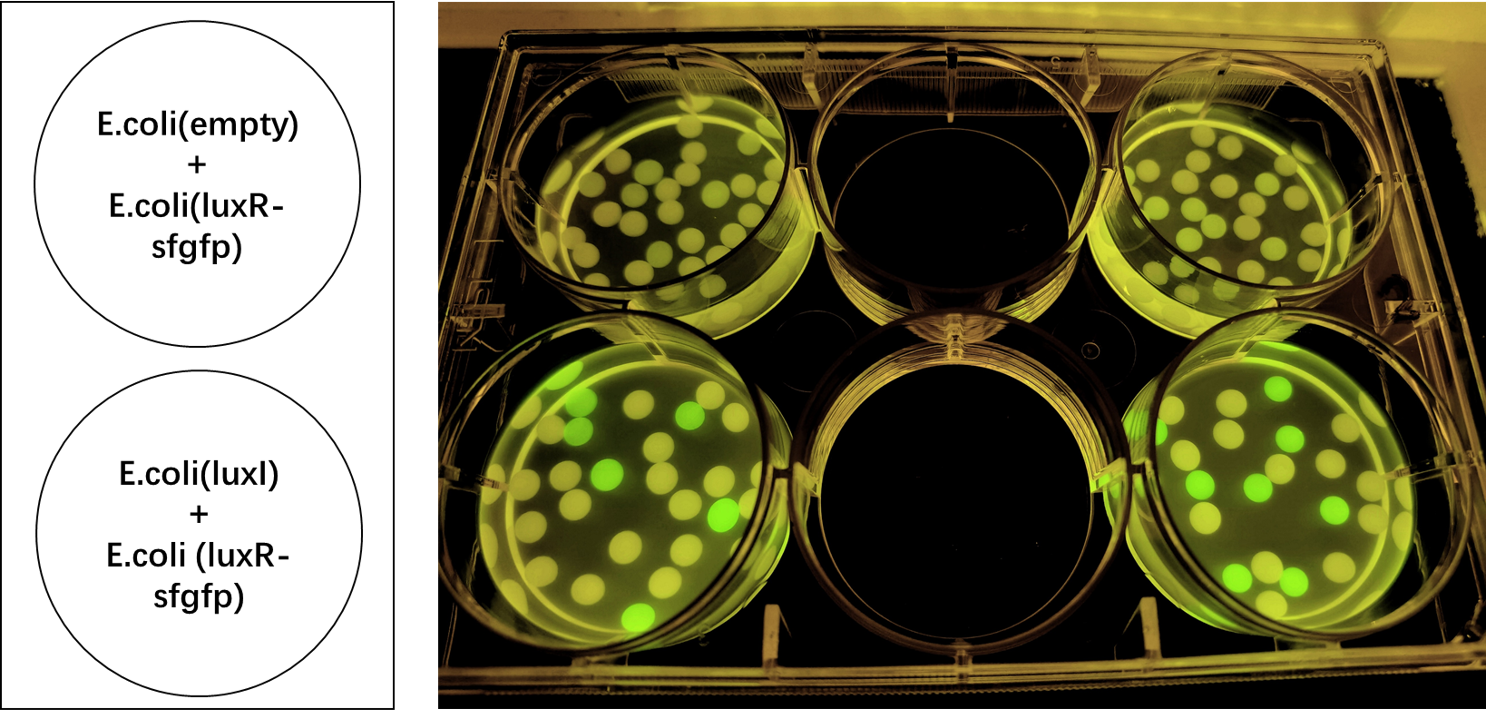
We also induced the expression of LuxR in sodium alginate gel beads and detected the expression of sfGFP under UV light. (Figure 3)
More detailed information can be seen on the composite page BBa_K4115039 , which is submitted by ShanghaiTech_China 2022.
Alma 2022 - Enzymatic and Structural Information on LuxI
LuxI, a acyl-homoserine lactone synthease (EC number), is responsible for creating the ‘auto-inducer’ molecule 3OC6-HSL. This molecule can be used to activate quorum sensing transcription factors such as LuxR, activating or repressing transcription in response to cell density (since AHLs and HSLs are diffusible outside the cell).
We wanted to design a kill-switch based on a quorum sensing module, and chose LuxI for this purpose. We wanted to learn more about this interesting protein in order to better model and design our genetic circuit.
First, we looked at different databases for information regarding LuxI and 3OC6-HSL. We found out that:
- In Vibrio Fischeri, 3OC6-HSL is synthesized at a rate of 3,300nmol/hr, or 0.91nmol/s. (BIONUMB3R5s database, entry 112005)
- AHL molecules are stable. 3OC6-AHL breaks down at a rate of 0.0003 per min, or 0.000005 per second. (BIONUMB3R5s database, entry 111994).
- LuxI is part of a family of auto-inducer syntheases, with an Enzyme Classification number of 2.3.1.184 (See the KEGG entry here: https://www.genome.jp/entry/2.3.1.184)
- LuxI requires SAM and an acylated acyl-carrier protein (ACP). However, LuxI is highly specific for hexanoyl-ACP, which partly explains why it gives rise to a specific product. The Km for these substrates are 0.96uM for hexanoyl-ACP and 130uM for SAM. Activity is maximal between 20 and 30C, and decreases to half at 18C or to 10% at 37C. At optimal conditions, the enzyme can produce 967pmol of product per minute, for each mg of enzyme. (Schaefer et al, 1996, Generation of cell-to-cell signals in quorum sensing: Acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein -https://www.pnas.org/doi/pdf/10.1073/pnas.93.18.9505)
- A similar protein, BjaI, has been crystalized and the kinetics have been studied. For that enzyme, it was found that it had a Kcat of 0.045mol/s and a catalytic efficiency of 21,430 per mol second. (Dong et al, 2017, Molecular basis for the substrate specificity of quorum signal synthases - https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5576808/)
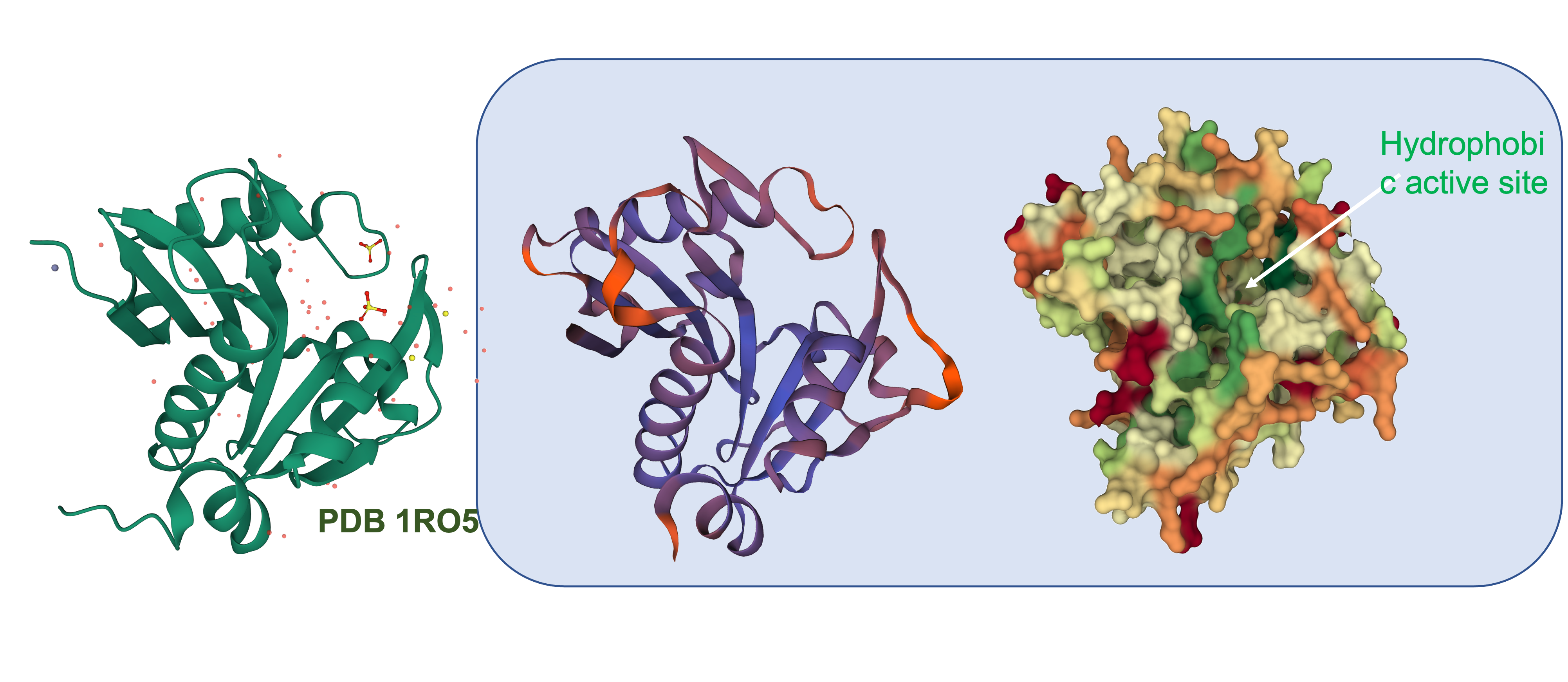
To get a better idea of how similar this enzyme is to other AHL synthases, we used the translated sequence and created a homology model using SWISS-MODEL.
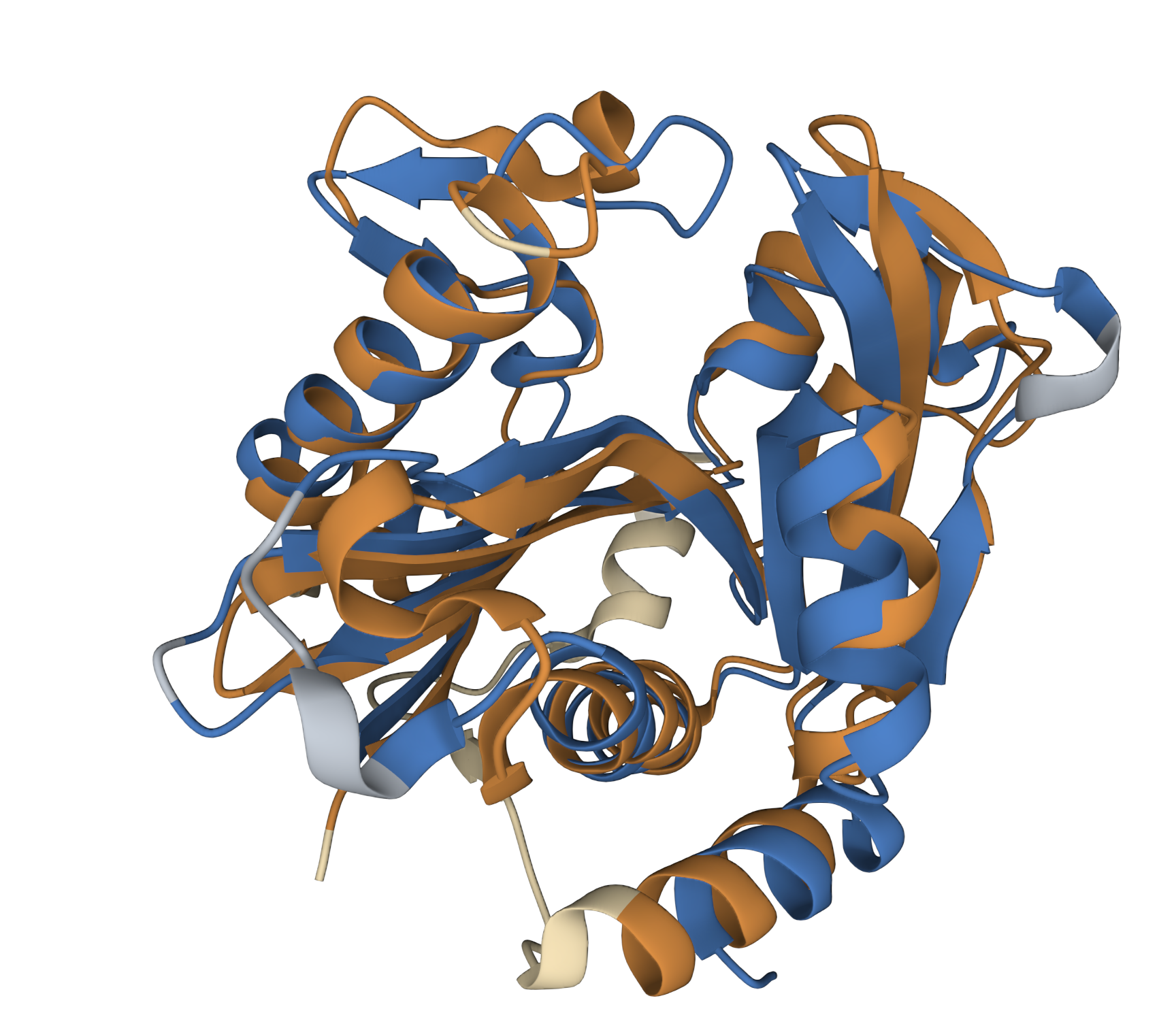
This model was based upon the template PDB structure “1RO5”, which is the LasI AHL synthase (similar, but produces a different type of AHL molecule). There was a 31% match between the primary sequence of LasI and LuxI. As a way of testing this model, and to make more direct structural comparisons, we took the generated structure of LuxI and did a pairwise structural alignment against the BjaI enzyme, which also produces the same AHL molecule as LuxI.
We found pretty close agreement between the two structures – both our LuxI model (Blue) and the BjaI structure, PDB code “5W8G” (Brown). There was an RMSD of 2.46, which is fairly good for structural comparisons or determination of structures (on average, the structure is displaced only 0.2nm). The alignment between the primary sequences can be seen below:
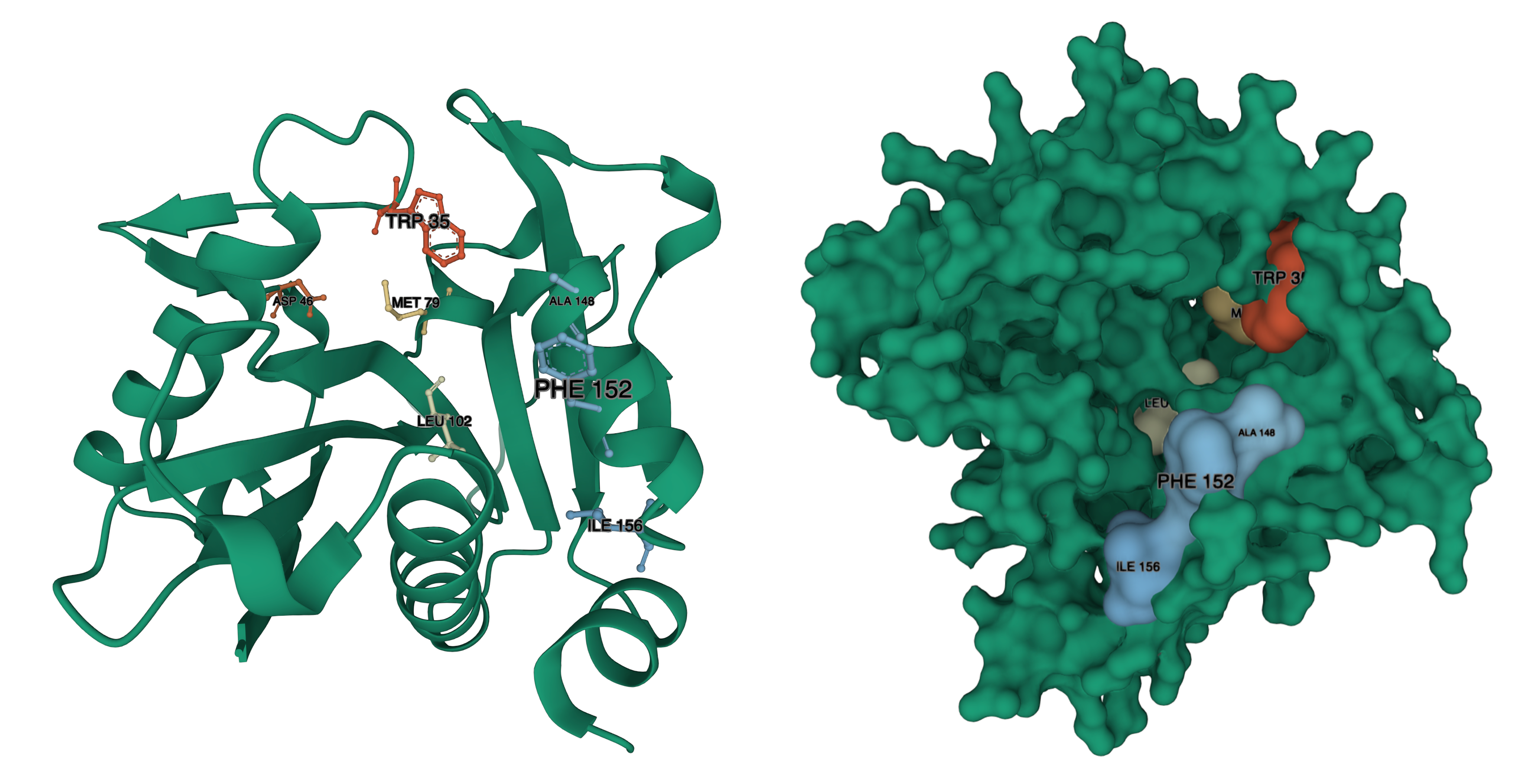
In the paper on BjaI (Dong et al 2017), a number of Alanine substitutions were explored that drastically decreased the Kcat. Others decreased or increased Km.
Decreases to Km, were observed with residues W34, M78, W101. Increases to Km, were observed with D46, W142, W143, F147. Increased to Km were also observed for Phenylalanine substitutions to W101, W142, and W143. We have identified where those residues are in our structural model:
BjaI residue – LuxI residue, from Red to Blue (Lower to Higher number): W34 – W35, D46 – D46, M78 – M79, W101 – L102, W142 – A148, W143 – F152 (there is a gap in the alignment, where BjaI has WW, LuxI has AIERF) F147-I156.
We hope that this information will assist teams that are planning on using LuxI in their experiments, or looking to modify LuxI activity by mutations.//cds/transcriptionalregulator/activator
//cds/transcriptionalregulator/repressor
//function/cellsignalling
//function/regulation/transcriptional
| direction | Forward |
| function | Mixed |
| ligands | HSL |
| protein | LuxR |
| swisspro | P12746 |
| tag | None |

 1 Registry Star
1 Registry Star