Part:BBa_K3799057
Bidirectional AIP-1 sensor
This is an improvement of the existing part BBa_K1022100.
Usage and Biology
This is a high throughput sensing module that can sense the presence of AIP-I molecules. This part consists of a pBAD promoter, an AIP sensor infrastructure (BBa_I746101), an AIP inducible promoter P2 and a GFP reporter.
In the natural system, the signalling Auto-inducing peptide (termed AIP) is made from AgrD, while AgrB, a transmembrane protein that is responsible for the cyclization. AgrC and AgrA, form the two-component system responsible for AIP detection and response relay, respectively. Upon binding to AIP, AgrC, a transmembrane histidine kinase receptor, facilitates the phosphorylation of the transcriptional activator AgrA. The phosphorylated AgrA, in turn, activates gene expression regulated by P2 and P3 promoters. There are four known variants of AIP (AIP I-IV) with different molecular structures and cross-inhibitory activity.
This part has been improved from BBa_K1022100 by flipping the direction of p2 promoter to the opposite direction of pBAD promoter.
In the existing part (Unidirectional circuit), we observed a significant increase in GFP expression in samples with zero AIP concentration. Based on this observation, we inferred that this could be due to the leaky gene expression of GFP. We have improved the existing part in such a way that the leaky gene expression reduced significantly.
Design
To solve the leaky gene expression we designed a bidirectional circuit. We Used Hi-Fi DNA assembly kit to make this bidirectional circuit. First, we divided the existing part (BBa_K1022100) into three segments and ordered those from Twist Bioscience. These three fragments were assembled in two different ways to produce the existing part as well as our designed improved part. For these assemblies, we designed two different sets of primers (a total of eight primers) using the NEBilder tool. We extended our three gene fragments with appropriate primers to produce overlap sequences at both ends of each fragment.
To make the improved part, we designed the overlap sequences in such a way that the p2 promoter along with the GFP reporter is flipped. This will make the direction of pBAD promoter and P2 promoter opposite to each other, which will significantly decrease the leaky gene expression of the GFP reporter.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1105
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1493
Illegal BamHI site found at 1045
Illegal BamHI site found at 2377 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 295
Illegal AgeI site found at 720 - 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
Charectarization
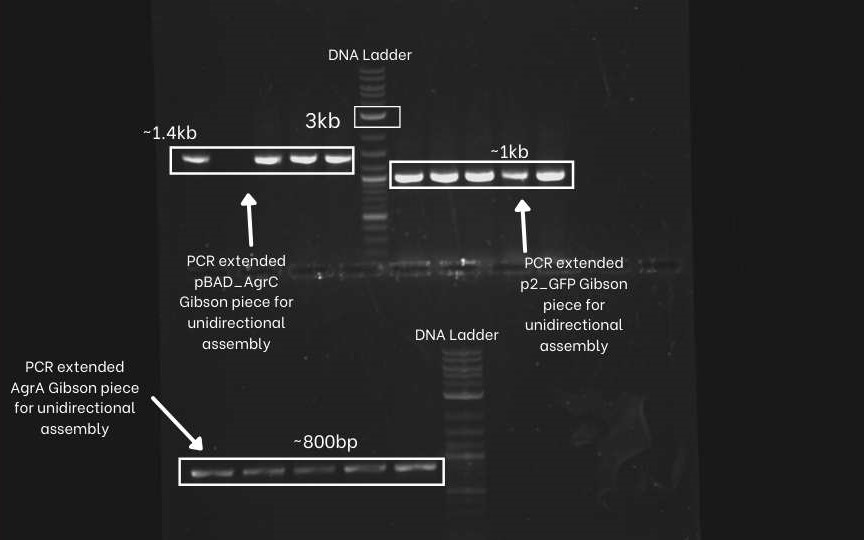
First, we PCR extend the three segments (pBAD-agrC, P2-GFP,agrA,) of the unidirectional circuit with specific primers. We did an Agarose Gel Electrophoresis to confirm if the Pcr extension was successful or not. As we can see below picture that we got the correct amplified bands for all three segments. (Extended pBAD-agrC ~ 1.4 Kb, Extended P2-GFP ~ 1Kb, Extended agrA ~ 800bp and extended backbone~2kb.
Next, We extend the same three segments (pBAD-agrC, P2-GFP, agrA and psb1C3 plasmid backbone) with different sets of specific primers. This extended overlap helps flip the P2-GFP fragments. We can see the gel electrophoresis image below. we can observe similar band sizes as above because the difference in length of the extension is very low.
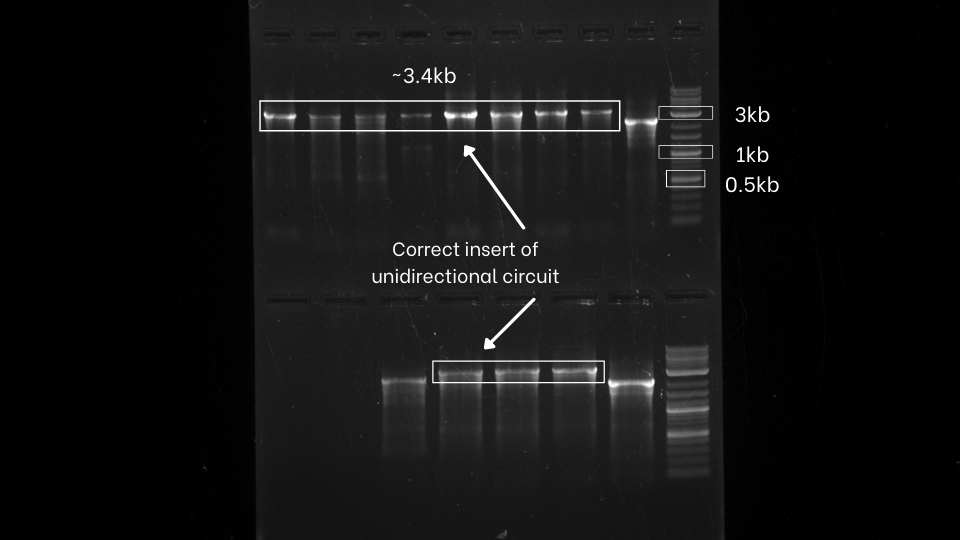
After the successful extension of the segments, we assemble those extended fragments using Hi-Fi DNA assembly master mix. then we directly transform that mix. After that, we perform colony PCR of both the unidirectional and bidirectional circuit using VF2 and VR to check whether we perfectly assemble the gene fragments.
From colony PCR result, We can observe that we got positive inserts for both unidirectional and bidirectional assemblies.
After the successful assembling of both the fragments, we planned to see the expression level of GFP in both bidirectional and unidirectional circuits. For that, we first express the AIP-1 generator (BBa_K3799006) in 40ml of bacterial culture to obtain AIP-1 molecules. After expressing for 8-10 hours, We centrifuge the culture and filtered sterilize the supernatant. To concentrate AIP-1 molecules, We further lyophilized the supernatant. After full lyophilization, we resuspended it in 3 ml of NFW to make a stock solution of AIP1.
We diluted our stock solution of AIP-1 molecules to make solutions of different concentrations, from 0% to 100%, where 0% is the negative control and 100% is the one-third dilution of our stock solution.
We induced our two types of cells (one with unidirectional plasmid and the other with bidirectional plasmid) with different concentrations of AIP-1 solution that we mentioned above. Then we incubated these two types of cells at 30 degrees centigrade for 7 hours.
We measured the fluorescence intensity of those two cells and observed that in absence of AIP-1 molecule, there is a lecky gene
| None |