Part:BBa_K3584002
T7 pro-tag-Lac ope-PPO-T7 ter
BBa_K3584002 is used to express Polyphenol oxidases(PPD) protein for Ni-sepharose purification.
Contribution
BBa_K3584000 is the coding sequence of a polyphenol oxidase (PPO) from Pleurotus ostreatus, which is different from the present enzymes showing the similar activity. PPO is a group of enzyme that are able to oxidize phenol or polyphenol by molecular oxygen to form the corresponding quinone. In a broad sense, polyphenol oxidase can be divided into three categories: monophenol monooxidase (tyrosinase, EC.1.14.18.1), bisphenol oxidase (catechol oxidase, EC. 1.10.3.2) and laccase (laccase, EC.1.10.3.1). Among these three types of polyphenol oxidase, catechol oxidase is mainly distributed in plants, and the polyphenol oxidase in microorganisms mainly includes laccase and tyrosinase. The polyphenol oxidase mentioned in most of the literature is generally the collective name of catechol oxidase and laccase.
Engineering Success
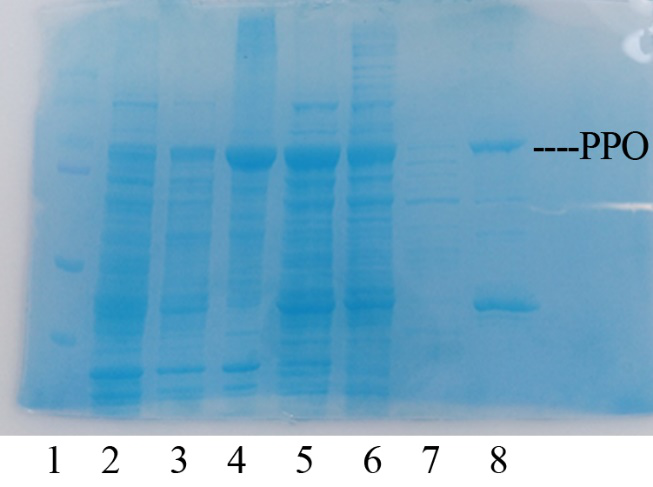
Production, purification, and SDS-PAGE analysis of recombinant PPO In order to present the function of the part, the PPO gene was expressed in E. coli BL21 (DE3) under the control of T7 promoter. Then the bacterial cells are collected and crushed, and the PPO enzyme solution is purified by further confirmation by the SDS-PAGE method, which is found in the corresponding protein band of approximately 57 kDa (Figure 1).
Figure 1. Production, purification, and SDS-PAGE analysis of recombinant PPO. Lane: protein molecular weight standard; lane 8: purified PPO.
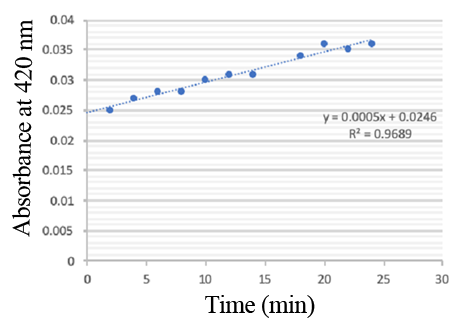
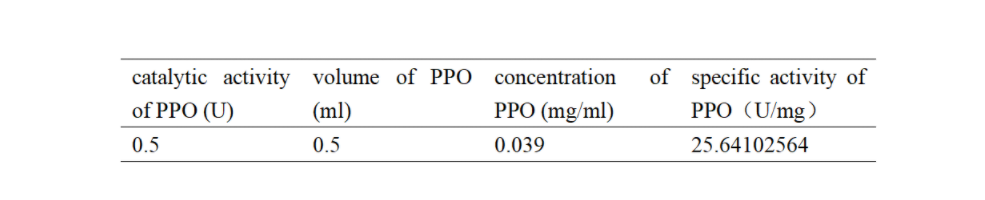
Enzymatic activity of the PPO PPO activity was determined using an analogue of p-cresol (catechol) as a substrate. In proper reaction mixture, PPO and 10 mM catechol was incubated at 50 ℃, pH 7.0 for 3 min, and the change in absorbance at 420 nm was measured spectrophotometrically. One unit of enzyme (PPO) activity was defined as the amount of enzyme that increased absorbance of 0.001 per minute. We found that as the extension of reaction time, reaction liquid of absorbance at 420 nm increased linear (Figure 2), we obtained by catalytic activity of PPO enzyme fluid, and divided by the corresponding protein concentration, the specific activity of PPO (Table 1).
Figure 2. PPO activity was determined using catechol as a substrate. The reaction mixture contained PPO protein and 10 mM catechol which was incubated at 50 ℃, pH 7.0 for 3 min, and the change in absorbance at 420 nm was recorded.
Table 1. Specific activity of recombinant PPO
Characterization by 2021iGEM_Shanghai_Metro_HS
Improvement of an existing part
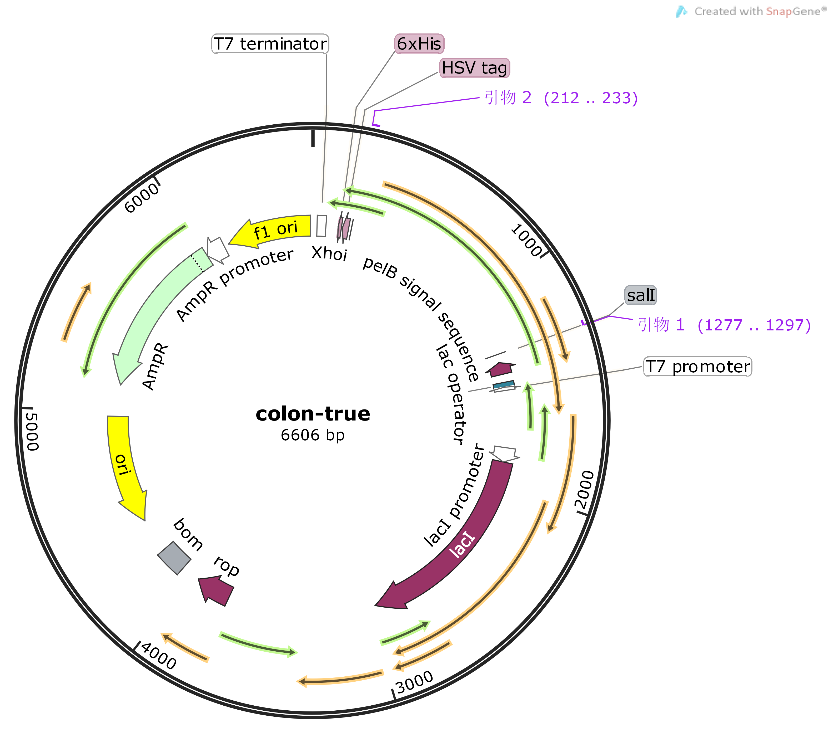
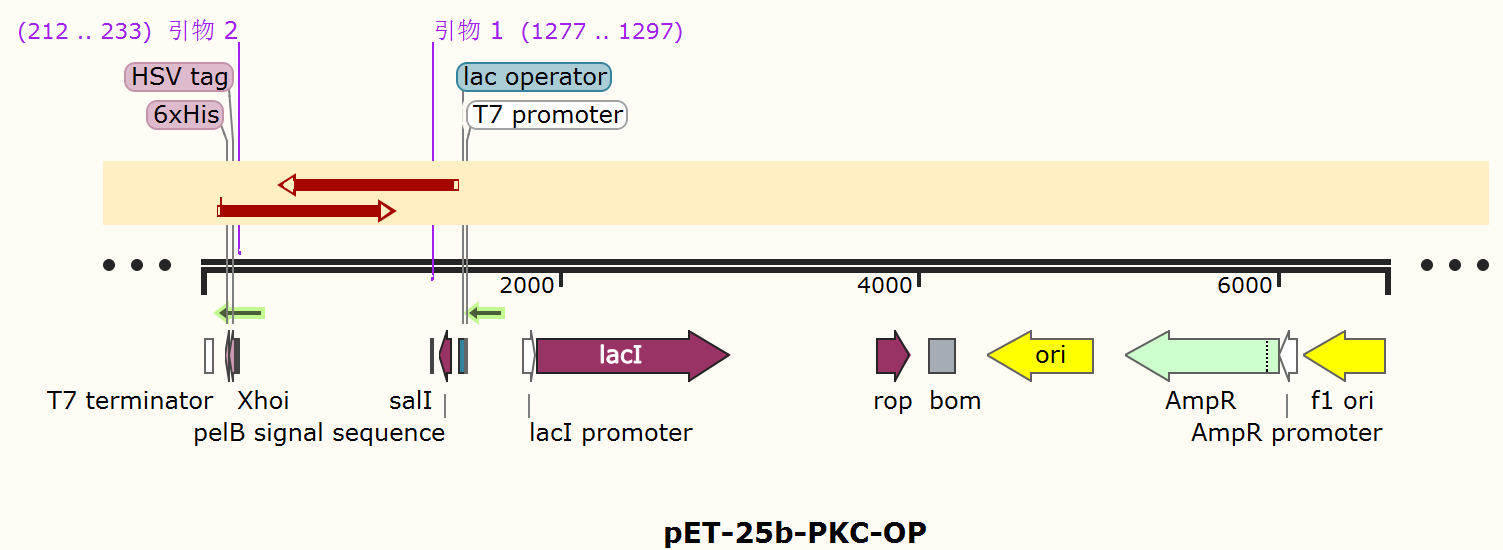
Compared to the old part BBa_K3584002, expressing T7 pro-tag-Lac ope-PPO-T7 ter, we design a new part BBa_K4096004, expressing T7 pro-Lac opePKC-OP-tag-T7 ter, with its sequence different from the old part (Figure 9). The group iGEM20_Shanghai_United aimed to degrade p-cresol sulfate for giving a potential treatment to chronic kidney disease patients. And they successfully constructed a composite part BBa_K3584002 and transformed it into E.coli BL21(DE3). They successfully expressed and purified PPO protein, and detected the enzyme activity. Differently, our team designed a new part BBa_K4096004 and aimed to construct an E.coli strain that can expressed alkali cellulase so as to improve ruminants' ability to acquire monosaccharides.. We chose T7 pro-Lac ope-PKC-OP-tag-T7 ter., which sequence is different from part BBa_K3584002 (Fig8), to construct our desired engineering strain.
Furthermore, in order to improve the utilization rate of silage and grain mixed feed, and the applicable varieties of poultry and ruminants. We constructed an improved composite part BBa_K4096006, which further add the Bacillus subtilis xylanase xynA gene to the cloning recombinant vector to make it compatible with cellulase PKC-OP and express β-xylosidase at the same time.
Profile
Name: pET-25b-PKC-OP
Base Pairs: 6633 bp
Origin: Synthetic
Properties: A recombinant plasmid containing cellulase sequence.
Usage and Biology
BBa_K4096004 is a plasmid that can expressed cellulase (PKC-OP) under the control of T7 promoter from Pseudomonas aeruginosa. Cellulases break down the cellulose molecule into monosaccharides ("simple sugars") such as beta-glucose, or shorter polysaccharides and oligosaccharides. Several different kinds of cellulases are known, which differ structurally and mechanistically. Synonyms, derivatives, and specific enzymes associated with the name "cellulase" include endo-1,4-beta-D-glucanase (beta-1,4-glucanase, beta-1,4-endoglucan hydrolase, endoglucanase D, 1,4-(1,3,1,4)-beta-D-glucan 4-glucanohydrolase), carboxymethyl cellulase (CMCase), avicelase, celludextrinase, cellulase A, cellulosin AP, alkali cellulase, cellulase A 3, 9.5 cellulase, and pancellase SS.
Construct design
The alkaline cellulase gene from Pseudomonas aeruginosa PKC-OP performed codon optimization on PKC-001 was selected and inserted into the pET-25b vector which contains a pelB signal peptide (Figure 1 and 2). The recombinant plasmid then was transformed it to E. coli BL21(DE3) strain to produce cellulase.
The profiles of every basic part are as follows:
BBa_K4096002
Name: PKC-OP
Base Pairs: 1086bp
Origin: Pseudomonas aeruginosa, genome
Properties: A coding sequence of alkali cellulase.
Usage and Biology
BBa_K4096002 is a coding sequence of alkali cellulase (PKC-OP) from Pseudomonas aeruginosa. Cellulases break down the cellulose molecule into monosaccharides ("simple sugars") such as beta-glucose, or shorter polysaccharides and oligosaccharides. Several different kinds of cellulases are known, which differ structurally and mechanistically.
BBa_K4096000
Name: pET-25b-vector
Base Pairs: 5547bp
Origin: Addgene
Properties: A plasmid expressing proteins.
Usage and Biology
BBa_K4096000 is a plasmid that can express proteins.
Experimental approach
Construction of recombinant plasmid
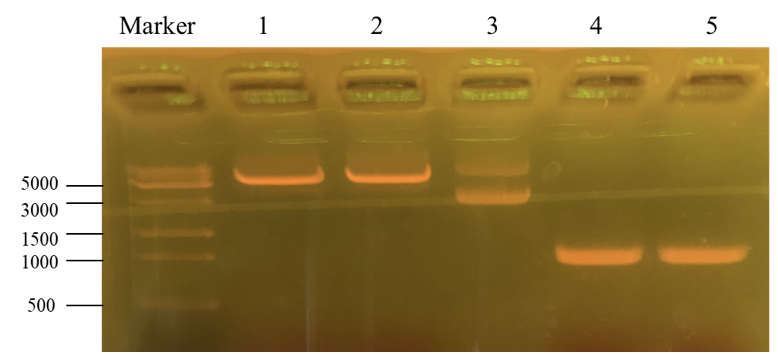
This picture is the nucleic acid electrophoresis result of enzyme cutting and PCR (By performing PCR, we can obtain the desirable PKC-OP’s gene segment.). Column “marker” is a column that is used to show the position of different lengths of genes. In this step, our aim is to verify whether these results are the desired ones. Number 1 and 2 is the result for pET-25b after enzyme digestion. Additionally, number 3 is the pET-25b without enzyme cutting. At last, number 4 and 5 is the PKC-OP after PCR. As our experiment moves on, the concentration of our PKC-OP and pET-25b are 233.2ng/μL and 17.8ng/μL. Lastly, we use the homologous combination to combine them.
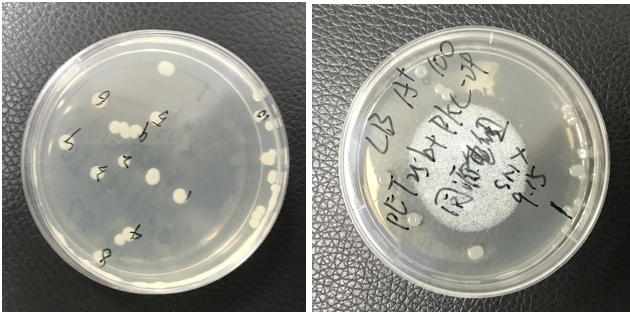
The pET25b-PKC-OP was constructed. The plate shows monoclonals of pET25b-PKC-OP constructs.
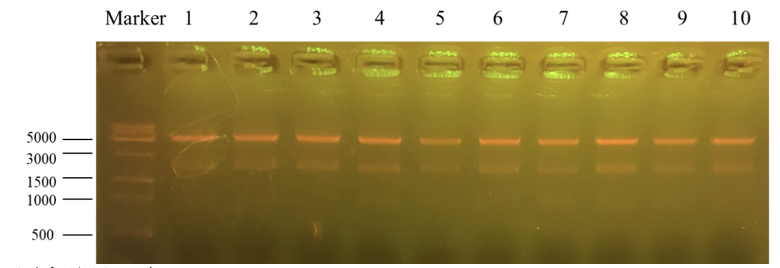
Column “marker” is a column that is used to show the position of different lengths of genes. Number 1 to 10 is the result for recombinant plasmid pET25b-PKC-OP after Apa1 enzyme digestion. We get two bands of 4712bp and 1894bp. It further indicates that the obtained monoclonals were positive monoclonals containing the recombinant plasmid. 1, 7 and 8 plasmids were sent to sequence.
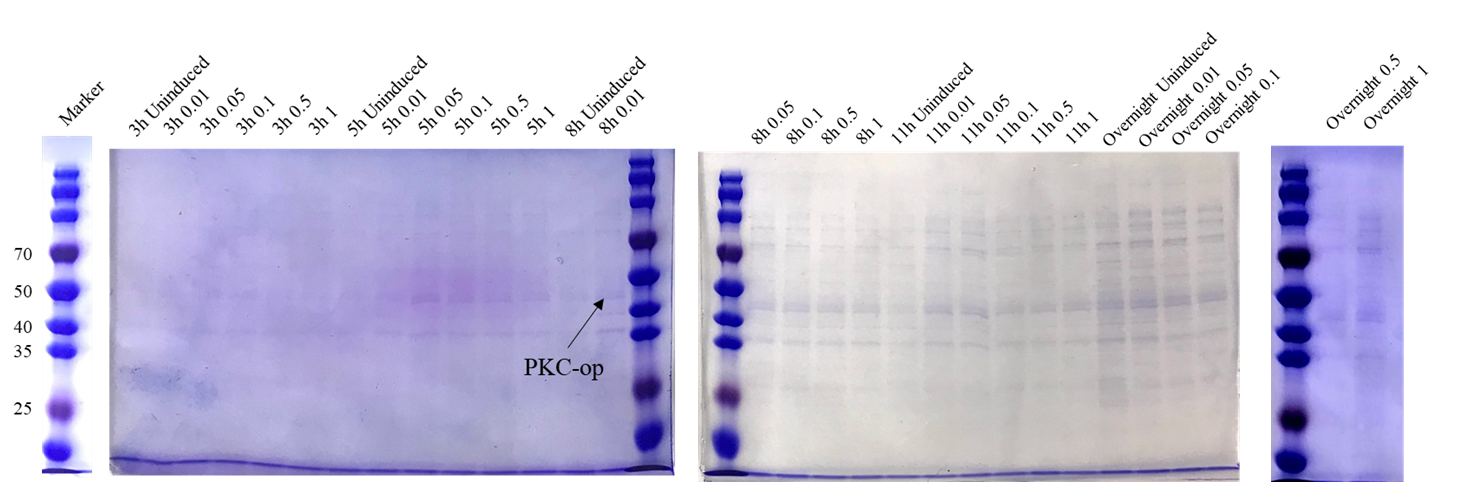
Sequencing feedback shows we have obtained the correct plasmids which is consistent with their DNA profiles. Different induction conditions were tested for protein expression. As the pET-25b vector contains a pelB signal peptide, the engineered strain would secret the protein into the medium. Therefore, we collected the culture supernatant after induction and ran SDS-PAGE for verification (Figure 2).
The theoretical molecular weight of the PKC cellulase is 45.6 kDa. As seen from the SDS-PAGE (Figure 7), there is a wide band just above 40 kDa and it indicates that we have obtained the PKC cellulase.
Proof of function
As our goal is to produce the additive that could secret PKC enzyme to help degrade the cellulose in silage, we want to ensure that our engineered strain can make the best of itself at its best condition. From this perspective, we designed several different induction conditions for protein expression and decided to build the model to determine the optimal condition for our engineered strain to “work” better.
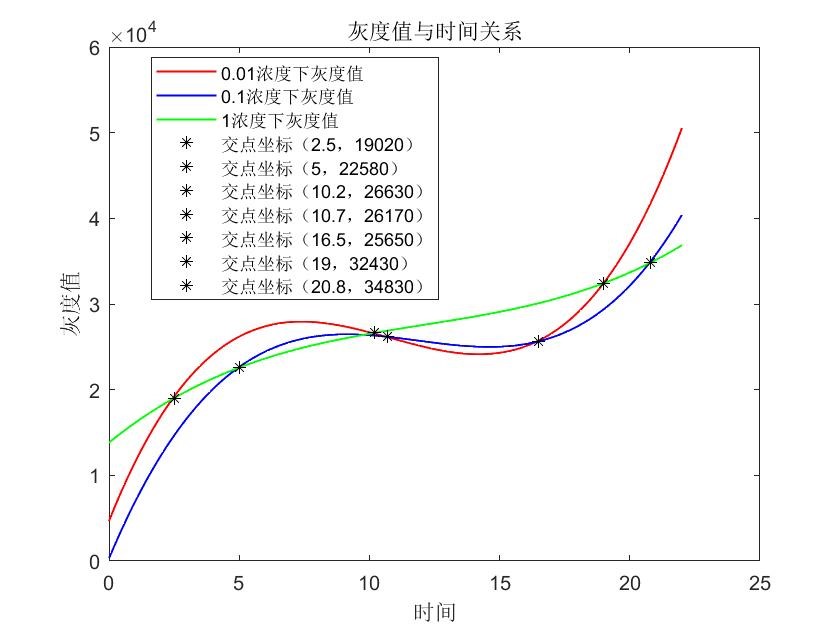
In table 1, gray values were measured by Image J in order to quantify the protein expression level. In this case, we tested three concentrations of IPTG to conduct induction. According to the scatter plots, we noticed that the cubic polynomial equation could fit the trends very well with all fitting degrees higher than 0.98.
Based on the graph where we can see several cross points’ coordinates of lines, we could adjust the IPTG concentration according to the induction time which could be pre-decided by the production plan in the future. 1.When the induction time is given less than 2.5 hours, 1 mM IPTG would be more recommended; 2.When the induction time is given during 2.5 hours 10.2 hours, 0.01 mM IPTG would be more recommended; 3.When the induction time is given during 10.2 hours ~ 19 hours, 1 mM IPTG would be more recommended; 4.When the induction time is given very enough like more than 19 hours, 0.01 mM IPTG would be more recommended.
Future plan
Testing the effectiveness of producing cellulase of positive recombinant bacteria. This is important because we need to know how much bacteria is needed to be used to effectively help animals digest fodder. In this experiment, we can control the time that positive recombinant bacteria grow and use the OD test to set the numbers of bacteria. Then put different numbers of bacteria and cellulose filter paper together. OD test can be used again to see the glucose concentration.
References
MUCK R,NADEAU E,MCALLISTER T,et al. Silage review: recent advances and future uses of silage additives [J]. J Dairy Sci,
刘海燕,张鹏举,王秀飞,等. 正交试验法优化玉米秸秆穰叶青贮发酵剂的研究 [J]. 中国饲料,2018 ( 11) : 74-79.
Coward-Kelly G., Aiello-Mazzari C., Kim S., Granda C., and Holtzapple M., 2003, Suggested improvements to the standard filter paper assay used to measure cellulase activity,Biotechnology & Bioengineering, 82(6): 745-749.
Ghose T.K., 1987, Measurement of cellulase activities, Pure & Appl Chem, 59(2): 257-268.
Lu Z.L., Chen D., Zhang S.S., Lu Q., and Huang R.B., 2012, A Pseudomonas aeruginosa producing alkaline cellulase, China Patent, 2012103821716
Characterization by 2021iGEM_Shanghai_HS_United
Improvement of an existing part
1.Both of the research focus on promoting the degradation of metabolites in the body by building engineering bacteria. Compared to the old part BBa_K3584002, they aimed to engineer probiotics that can be consumed by the patient, express the target gene into enzymes to degrade p-cresol, the precursor of pCS. In our project, we put FLR gene into E. coli to produce a strain secreting FLR enzyme. The strain degrades flavonoids to produce DAT and stimulate the immune system to achieve the purposes of anti-inflammatory, antibacterial and anti-cancer. Both of our research objectives are to promote the degradation of metabolites in the body by building engineering bacteria.
2. About plasmid construction, our improvement is reflected in the construction of recombinant plasmids that express different functions using the same promoter-terminator model, composite part T7 pro-tag-Lac ope-XX-T7 ter.
Compared to the old part BBa_K3584002, composite part T7 pro-tag-Lac ope-PPO-T7 ter., we design a new part BBa_K3998000 which replaced the PPO fragment with the Flr gene fragment. Flr enzyme was successfully produced by transformed Escherichia coli. On the basis of purified recombinant Flr, an excellent Flr activator was obtained. After we obtained the purified protein Flr, namely the FLR enzyme, we conducted enzyme activity tests by using 4 kinds of flavonoids samples: apigenin, chrysin, luteolin, diosmetin with the initial concentration 10mg/L and the concentration of FLR enzyme was 1mM/L. Each sample was guaranteed three replicates of the enzyme activity test in order to gain more data to ensure the credibility of the result of our experiment. All these results indicate that our enzyme has high activity and is capable of degrading multiple flavonoids with a certain universality.
flr
Profile
Name: flr
Base Pairs: 6275bp
Origin: Synthesis from a genetic company
Properties: A protein used to improve the degradation of flavonoids
Usage and Biology
In this project, we put the FLR gene into E. coli to produce a strain secreting FLR enzyme efficiently. This strain can better express the FLR gene, improve the degradation of flavonoids, further produce DAT and stimulate the immune system of the human body, so as to achieve anti-inflammatory, antibacterial, anti-cancer and other purposes, at the same time to help reducing clinical treatment costs.
Construct design
The target protein expression fragment(BBa_K3998000) is constructed into a vector of pET28a. According to Yang, G., et al. (2021), flavone reductase (FLR) discovered from Flavonifractor plautii ATCC 49531 (originally assigned as Clostridium orbiscindens DSM 6740) plays a key step in catalyzing flavonoid. Thus, we plan to over-express FLR in Eco. li (BL21) and test its function in degrading flavonoid. The composite part is transcripted by the T7 promoter and stopped by the T7 terminator. Meanwhile, a His protein tag is inserted for future protein purification. We carried out molecular biology experiments and successfully constructed the composite part above in the vector of Eco. Li (BL21).
Experimental approach
In lab, we successfully constructed the plasmid and it was proved by colony PCR and sequencing result.
Preparation of pET28a vector: The vector was obtained from our plasmid library.
Acquisition of Inserts: Introducing homologous sequences of pET28a vector into 5’-end of Forward (F) & Reverse (R) primer, respectively, aiming to make the ends of amplified inserts and vectors identical to each other.
Recombination: Calculated the amount of DNA for recombination by formula. Diluted pET28a vector and inserts before recombination to make sure the loading accuracy.
Transformation
Place the competent cells on ice (i.e. DH5α competent strain). 2.Pipet 10 μl of the recombination products to 100 μl of the competent cells, flip the tube several times to mix thoroughly (DO NOT VOTEX!), and then place the tube still on ice for 30 min. The volume of transformation products should not be more than 1/6 of the volume of competent cells. 3. Heat-shock the tube at 42℃ for 45 sec and then immediately chill on ice for 2 - 3 min. 4. Add 900 μl of LB medium (without antibiotics) to the tube. Then, shake at 37℃ for 1 hour at 200 - 250 rpm. 5. Preheat the LB plate which contains appropriate selection antibiotic at 37℃ . 6. Centrifuge the culture at 5,000 rpm for 5 min, discard 900 μl of supernatant. Then, re-suspend the pellet with 100 μl of remaining medium and plate it on an agar plate which contains appropriate selection antibiotic. 7. Incubate at 37℃ for 12 -16 hours. we successfully constructed the plasmid and it was proved by colony PCR and sequencing result.
Proof of function
Enzyme Activity Test of FLR
After obtaining the purified protein containing FLR enzyme, we tested its effectiveness in degrading flavonoid. We conducted enzyme activity tests by using 4 kinds of flavonoid samples: apigenin, chrysin, luteolin, diosmetin with the initial concentration 10mg/L and the concentration of FLR enzyme was 1mM/L. Each sample was guaranteed three replicates of the enzyme activity test in order to to ensure the credibility of the test results. The test results are listed as follows:
Results show that the concentration of these 4 flavonoids remarkably decreases after 2 hours which were degraded by the enzyme, even there were only 0.3-5% left after 6 hours. All these results indicate that our enzyme has high activity and is capable of degrading multiple flavonoids with a certain universality.
Bacteria Activity Test
All these flavonoids were almost degraded after 2 hours with only 1-7% left, which indicates the strong ability of our engineered E. coli to degrade flavonoids as we expected. Above all, we could come to the conclusion that the FLR enzyme, as well as the engineered E. coli, is capable of degrading various flavonoids with a certain universality and practicability. In comparison, the degradability of FLR enzyme to these flavonoids could be ranked from greatest to least as apigenin > chrysin > luteolin > diosmetin.
Future Plan
Since the FLR enzyme showed a great ability to decompose flavonoids, we are looking forward to testing how the human body would consume with and without the FLR enzyme. What’s more, if the further experiment succeeds, we will probably try to add the FLR engineered bacteria into yogurt. Yogurt contains a large number of lactic acid bacteria, which themselves belong to acidic substances. After use, they can promote gastrointestinal peristalsis and food digestion. Yogurt helps in digesting food and FLR helps in digesting flavonoids. By adding it, we could make a health-care yogurt which is great for human health improvement.
References
1. Gaohua Yang, Sen Hong, Pengjie Yang, et al. Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria, Nature communcations (2021).
2. 黄瑶. 黄酮类物质改善认知功能障碍作用机制的研究进展[J]. 养生大世界 2021年4期, 241-242页, 2021.
3. Thilakarathna S H, Rupasinghe HP. Flavonoids Bioavailability and attempts for bioavailability enhancement[J].Nutrients, 2013, 5(9):3367-3387
4. Ravishankar D, Rajora A K, Greco F, et al. Flavonoids as prospective compounds for anti-cancer therapy[J].Int J Bio- chem Cell Biol, 2013, 45(12):2821-2831
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 169
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1428
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 380
- 1000COMPATIBLE WITH RFC[1000]
| None |