Part:BBa_K2217001:Design
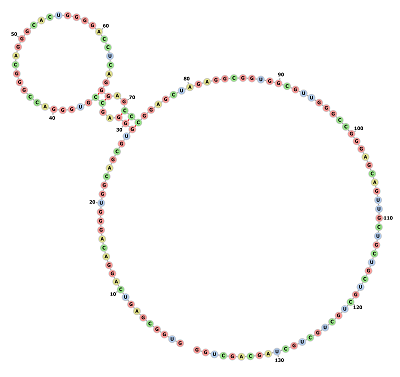
Circular RNA hsa-circ-000064
We have used Vienna RNA package[1] for generating structural models of circular RNA hsa_circ_0000064. RNA sequence was retrieved from circ-Interactome database[2] to be used as an input for Vienna package. Vienna RNA package depends on extension of linear folding algorithms. Circular RNA molecules are modelled through post-processing of computed linear arrays. Using Vienna RNA Package, we could compare structural modifications between linear and circular structures in a memory-effective manner.[3] The energy contribution of Exterior loop should be scored in circular structures, on the other hand, exterior loops have no energy contribution in linear structures. RNAfold structure prediction tool was used to calculate the minimum free energy (MFE) and backtraces an optimal secondary structure. To compute centroid structure we used McCaskill's algorithm[4] through -p option. Mountain plot was produced with mountain.pl, Dot plot was also generated by RNAplfold.
Design Notes
Minimum free energy prediction using RNAFOLD generated an optimal secondary structure in dot-bracket notation from a centroid structure of 0.00 kcal/mol minimum free energy to 1.78 kcal/mol. Thermodynamic ensemble prediction using RNAFOLD computed a free energy of -51.72 kcal/mol, The frequency of the MFE structure in the ensemble is 0.07 % and the ensemble diversity is 65.27.
The energy dot 2D plot which indicated all of the base pairs involved in optimal and suboptimal secondary structures, both axes of the graph represent the same RNA sequence. Each point drawn indicates a base pair between the ribonucleotides whose positions in the sequence are the coordinates of that point on the graph. The Mountain plot included the MFE structure, the thermodynamic ensemble of RNA structure, and the centroid structure. Additionally we used it to estimate the positional entropy for each position.
More Information about circular design of this part can be found at [http://2017.igem.org/Modeling.html# Egypt-AFCM Modeling]
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 128
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Source
Synthesised as a basic part in a Gene Fragment by IDT
References
[1] Lorenz, R. and Bernhart, S.H. and Höner zu Siederdissen, C. and Tafer, H. and Flamm, C. and Stadler, P.F. and Hofacker, I.L. "ViennaRNA Package 2.0", Algorithms for Molecular Biology, 6:1 page(s): 26, 2011
[2] Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biology. 2016;13(1):34-42. doi:10.1080/15476286.2015.1128065.
[3] Hofacker I L, Stadler P F. Memory efficient folding algorithms for circular RNA secondary structures.Bioinformatics, 2006, 22: 1172-1176
[4] McCaskill J. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119.