Part:BBa_R0062
Promoter (luxR & HSL regulated -- lux pR)
Promoter activated by LuxR in concert with HSL
The lux cassette of V. fischeri contains a left and a right promoter. The right promoter gives weak constitutive expression of downstream genes.This expression is up-regulated by the action of the LuxR activator protein complexed with the autoinducer, 3-oxo-hexanoyl-HSL. Two molecules of LuxR protein form a complex with two molecules of the signalling compound homoserine lactone (HSL). This complex binds to a palindromic site on the promoter, increasing the rate of transcription.
This Plux promoter "pointing to the right" is the same sequence, but inverted, as part BBa_K199052.
Usage and Biology
Pretty good off in the absence of LuxR/HSL. [jb, 5/24/04]
The team from Davidson College and Missouri Western State University discovered that this part promotes "backwards transcription" when LuxR protein is present and AHL-3OC6 is absent. You can read [http://www.ibc7.org/article/journal_v.php?sid=265 the paper that documents this unexpected "backwards promoter activity"] in their open access paper.
Tsinghua-A 2017's characterization
Crosstalk between Plux and Nine AHL-Receptor Combinations
I Background
The crosstalk of Plux between 3 kinds of AHL molecules (3OC6HSL, 3OC12HSL and C4HSL) and 3 kinds of receptors (LuxR, LasR and RhlR) was characterized by ETH_Zurich 2014. However, they used AHL reagents to measure the dose-response of promoter activity, which may not be convenient in quorum sensing, because the concentration of AHL bacteria secrete may be out of the range they tested before. In other words, bacteria is unable to secrete so much AHL molecules to activate gene expression. Furthermore, the response time in the quorum sensing process is also important for designers to optimize their synthetic circuits with better precise and robustness. Therefore, we designed circuits to measure the crosstalk between receptors expressed in one kind of E.coli and AHL molecules secreted by another kind of E.coli and response time of these systems.
II Results
Crosstalk between Plux and Nine AHL-Receptor Combinations

Fig. 1: Results of our crosstalk experiment
3OC6HSL-LuxR is the wild-type pair of Plux. However, from this result, we can see that there are many other pairs can activate Plux. For example, 3OC6HSL-LasR, 3OC6HSL-RhlR, 3OC12HSL-LuxR, C4HSL-LuxR, C4HSL-LasR, C4HSL-Rhl can all activate it. What is more, the quorum sensing needs more than 30 hours to establish a steady state, which may be important for designing dynamic functions in bacteria with quorum sensing.
III Experiment Design and Measurement Methods
First, we cloned three AHL synthases: luxI (BBa_C0061), lasI (BBa_C0078) and rhlI (BBa_C0070), each ligated with promoter (BBa_J23100), RBS (BBa_B0034) and terminator (BBa_B0015) on a low copy backbone (pSB3K3). At the same time, we cloned three receptor-promoter combinations to pSB6A1. The three receptors are luxR (BBa_C0062), lasR (BBa_C0179) and rhlR (BBa_C0171) (with the same promoter, RBS and terminator as AHL synthases). Sensor promoter Plux (BBa_R0062) drives RFP (BBa_E1010) expression (with the same RBS and terminator as above) to detect the response of the promoters to AHL molecules via the receptors. Adding together, there are nine AHL-Receptor-Plux combinations. After that, we co-transformed the plasmids containing AHL synthases and plasmids containing receptor-Plux combinations. (Gene circuit of them is shown in Fig. 2)

Fig. 2 Gene circuit of 9 AHL-Receptor-Plux combinations
Finally, we detect how RFP intensity per cell change with time, which can be used to indicate the intensity of the promoter response to AHL molecule via corresponding receptor. Detailed measurement methods are as follows:
1. Transform MG1655ΔsdiAΔlacI with 9 kinds of combinations of plasmids mentioned above.
2. Pick bacterial clones from the petri plate, then shake it overnight in the LB medium (3ml) with 50 microgram/ml Ampicillin and 30 microgram/ml Kanamycin at 37℃. For each combination, 2 clones are picked.
3. Dilute the overnight culture to 1/50 in fresh LB medium (3ml) containing 50 microgram/ml Ampicillin and 30 microgram/ml Kanamycin.
4. Incubate the fresh cultures at 37℃ until OD600 reaches 0.2.
5. Add 1ml culture obtained from Procedure 4 to 2ml fresh LB medium containing 50 microgram /ml Ampicillin and 30 microgram/ml Kanamycin. Incubate the fresh cultures at 37℃.
6. Take out 200μl cultures obtained from Procedure 5 and measure the fluorescent intensity and OD600 after some time. (The cultures will not be put back again.) The excitation wavelength is 584nm, emission wavelength is 607nm.
IV Data file of results shown above
TUST 2017's characterization
design of ultrasensitive responses basis of https://parts.igem.org/Part:BBa_R0062 use Point mutation
Background information
Quorum-sensing bacteria such as Vibrio fischeri, are able to detected their own population density and implement density-based decision-making,Using the luxI/luxR quorum-sensing system, synthetic biologists have designed a large number of devices in prokaryotic microorganisms.Inevitably, high performance is required from such devices, and this includes reliability, sensitivity, Hence, to improve the properties of population-density switches, we design of ultrasensitive responses base https://parts.igem.org/Part:BBa_R0062 use Point mutation
Experiment Design

We selected the successful strains of the experiment and sequenced part1: https://parts.igem.org/Part:BBa_K2267029 part3: https://parts.igem.org/Part:BBa_K2267030 part4: https://parts.igem.org/Part:BBa_K2267031 part6: https://parts.igem.org/Part:BBa_K2267032 part8: https://parts.igem.org/Part:BBa_K2267033
Methods
The experiments for the characterization of parts were performed as described previously For density-response testing, cells (E. coli strain BW25113) from single colonies on LB agar (BD, USA) plates were grown overnight in 1 ml nutrition-rich, acid-base equilibrium medium (REM) (15.2 g/l yeast extract (BD, USA), 0.5% (NH4)2SO4, 4 mM MgSO4, 2% glucose and 24 g/l K2HPO4.3H2O, and 9.6 g/l KH2PO4) in Falcon tubes overnight (8−12 h, 1000 rpm, 37 °C, mB100-40 Thermo Shaker, AOSHENG, China). The cultures were subsequently diluted 500-fold with REM in 96-well plates, which were further incubated at 37°C in a shaker at 1000 rpm. Once the diluted cultures reached an OD600 of 0.12–0.14 (~3 h), 10 μL aliquots were transferred into 1 mL REM in 24-well plates (Corning/Costar 3524). These plates were incubated at 37°C in a Varioskan Flash (Thermo Scientific, USA) under constant shaking at 1,000 rpm for 20 h to maintain exponential growth, during which the OD600 and fluorescence values were recorded
Tsinghua 2018's characterization
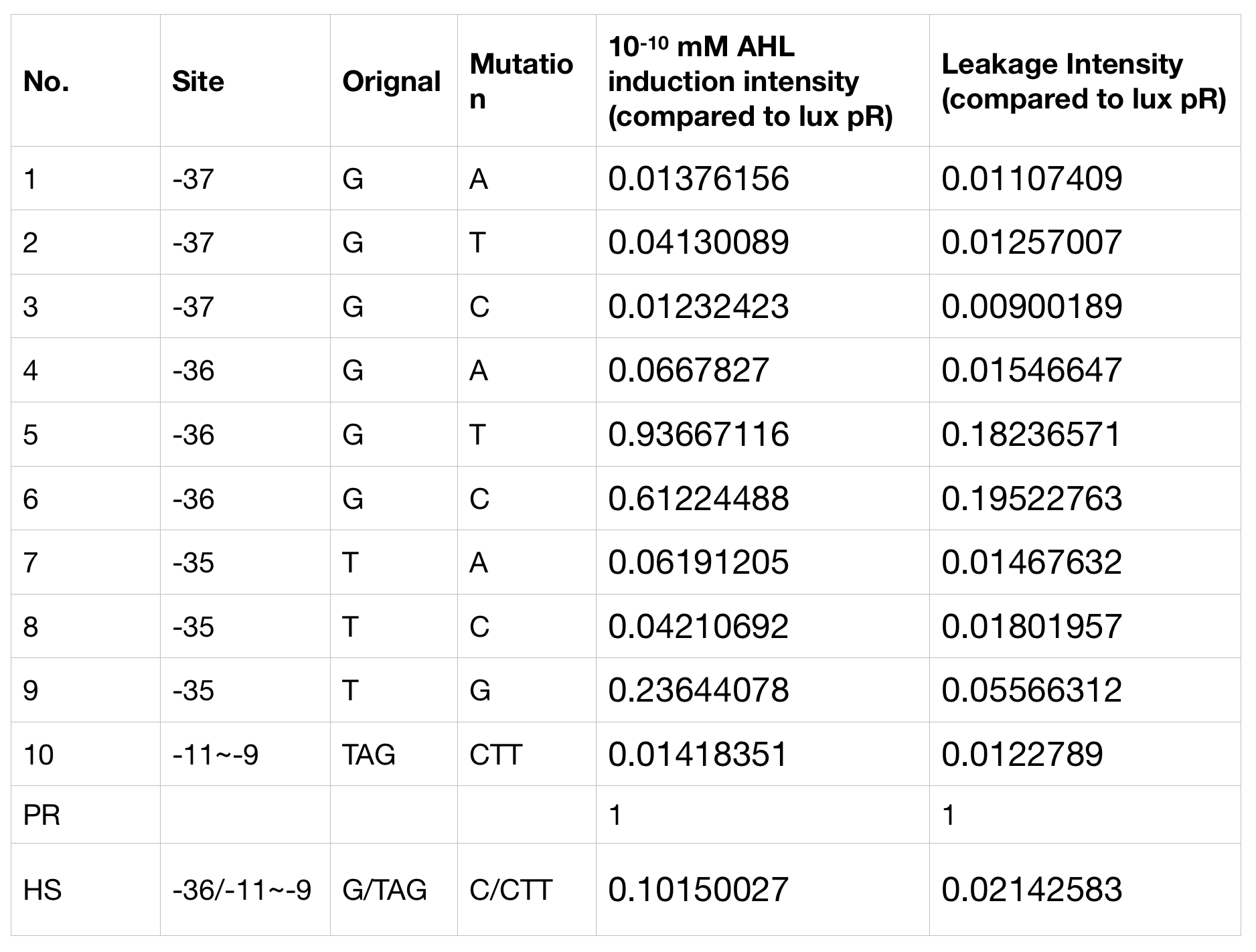
Lux pR Mutants in Different Gene Expression Strength, Leakage and Expression Perimeters
I Background
With the hope to find an optimal promotor for our NEON system, we designed 9 mutations on sites -35, -36, -37 near the luxR binding site and tested another on -10 site that TUST 2017 reported to show decreased leakage. We conducted experiments to evaluate these lux pR mutants’ reaction to AHL stimulation and their leakage level. The test devices we designed include a constantly expressed luxR and a lux pR (or mutant) driven sfGFP (like BBa_K2558211 with original lux pR promotor, and BBa_K2558212 with lux pR-HS promotor).
II Results
We conducted the experiment first with 9 mutations on sites -35, -36, -37 near the luxR binding and another on -10 site that TUST 2017 reported to show decreased leakage. We transferred the plasmids with lux pR driven sfGFP into E. coli DH5α. In theory, AHL can induce gene expression by activating lux pR promotor. We measured the value of Fluorescence/OD without AHL stimulation by microplate reader to show the leakages of different lux pR mutants, as lux pR promotor is not supposed to be activated without AHL. Meanwhile, we also regard the value of Fluorescence/OD with 10^-10 M AHL stimulation to observe the sensitivity and expression intensity of different lux pR mutants. Experiment data is shown in the figures below. (Figure.1)
-
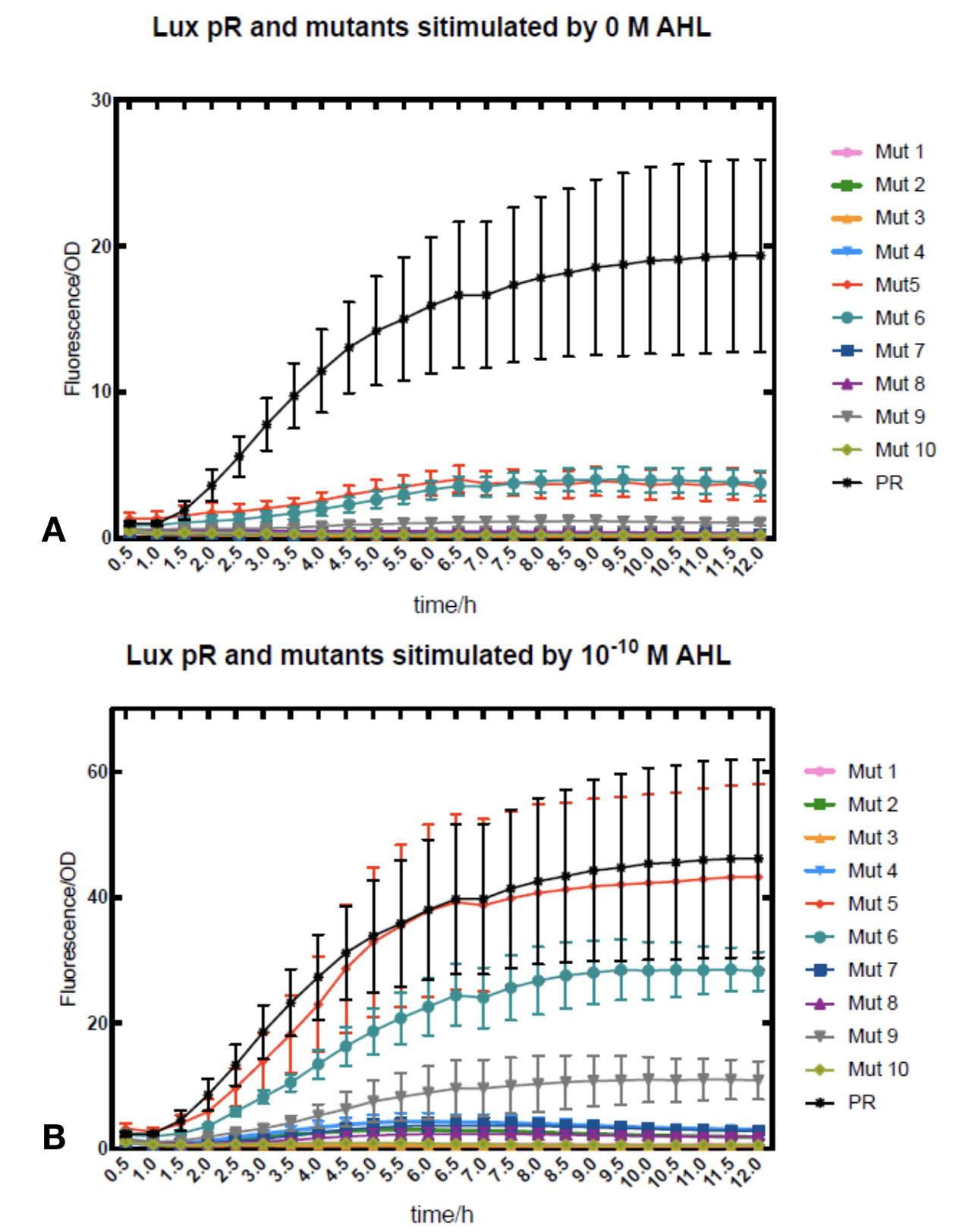 Figure.1. Leakage and 10^-10 mM AHL stimulation of lux pR promotor and its mutations. The fluorescence strengths and OD values are measured by microplate reader respectively at 510 nm and 600 nm wavelength. Plasmids with lux pR promotor and its mutants are transferred into E.coli DH5α. A The bacteria were treated without AHL inducing. High (Fluorescence/OD) ratio indicates strong leakage of promotor. Among all the strains, lux pR promotor has the strongest leakage and the largest error bar as well. B The bacteria were treated with 10-10 mM AHL induction. High (Fluorescence/OD) ratio indicates high expression intensity. Mutation 5 has almost the same expression intensity as the wild type lux pR promotor.
Figure.1. Leakage and 10^-10 mM AHL stimulation of lux pR promotor and its mutations. The fluorescence strengths and OD values are measured by microplate reader respectively at 510 nm and 600 nm wavelength. Plasmids with lux pR promotor and its mutants are transferred into E.coli DH5α. A The bacteria were treated without AHL inducing. High (Fluorescence/OD) ratio indicates strong leakage of promotor. Among all the strains, lux pR promotor has the strongest leakage and the largest error bar as well. B The bacteria were treated with 10-10 mM AHL induction. High (Fluorescence/OD) ratio indicates high expression intensity. Mutation 5 has almost the same expression intensity as the wild type lux pR promotor. -
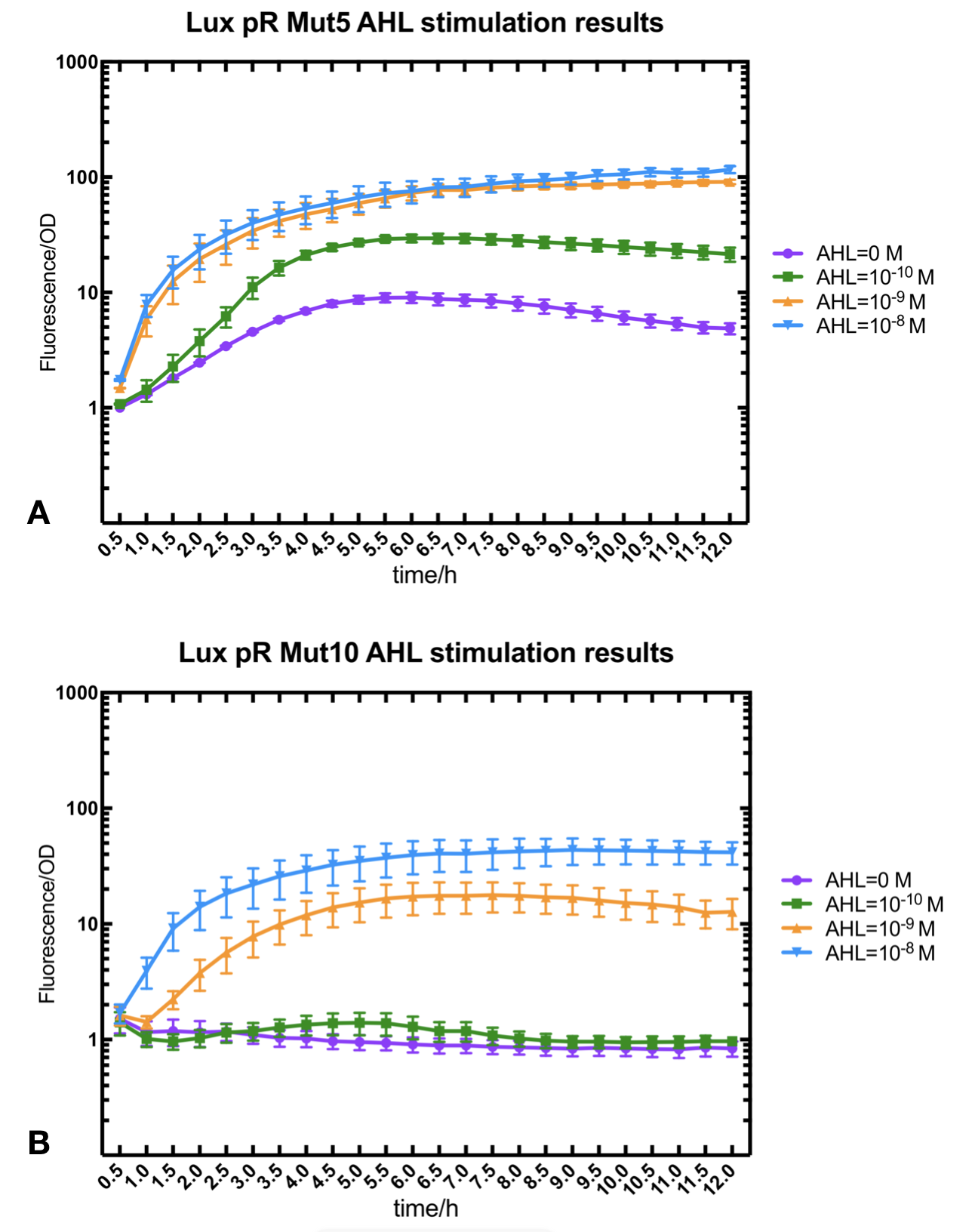 Figure.2. Leakage and AHL stimulation of of lux pR Mutant 5 and Mutant 10. The fluorescence strengths and OD values are measured by microplate reader respectively at 510 nm and 600 nm wavelength. A Plasmid with lux pR Mutant 5 is transferred into E.coli DH5α. The bacteria were treated without or with 10^-10 mM, 10^-9 mM and 10^-8 mM AHL inducing. B Plasmid with lux pR Mutant 10 is transferred into E.coli DH5α. The bacteria were treated without or with 10^-10 mM, 10^-9 mM and 10^-8 mM AHL inducing.
Figure.2. Leakage and AHL stimulation of of lux pR Mutant 5 and Mutant 10. The fluorescence strengths and OD values are measured by microplate reader respectively at 510 nm and 600 nm wavelength. A Plasmid with lux pR Mutant 5 is transferred into E.coli DH5α. The bacteria were treated without or with 10^-10 mM, 10^-9 mM and 10^-8 mM AHL inducing. B Plasmid with lux pR Mutant 10 is transferred into E.coli DH5α. The bacteria were treated without or with 10^-10 mM, 10^-9 mM and 10^-8 mM AHL inducing. -
 Figure.3. Leakage and 10^-10 mM AHL stimulation of of lux pR promotor WT, Mutant 5, Mutant 10 and HS. The fluorescence strengths and OD values are measured by microplate reader respectively at 510 nm and 600 nm wavelength. Plasmids with lux pR promotor WT, Mutant 5 and Mutant 10 are transferred into E.coli DH5α. A The bacteria were treated without AHL inducing. High (Fluorescence/OD) ratio indicates strong leakage of promotor. B The bacteria were treated with 10^-10 mM AHL induction. High (Fluorescence/OD) ratio indicates high expression intensity.
Figure.3. Leakage and 10^-10 mM AHL stimulation of of lux pR promotor WT, Mutant 5, Mutant 10 and HS. The fluorescence strengths and OD values are measured by microplate reader respectively at 510 nm and 600 nm wavelength. Plasmids with lux pR promotor WT, Mutant 5 and Mutant 10 are transferred into E.coli DH5α. A The bacteria were treated without AHL inducing. High (Fluorescence/OD) ratio indicates strong leakage of promotor. B The bacteria were treated with 10^-10 mM AHL induction. High (Fluorescence/OD) ratio indicates high expression intensity.We repeated AHL stimulation test on Mutant 5 and Mutant 10 (Figure.2) and decided to combine the two mutations together to form a new promotor with high intensity and small leakage. We named it lux pR-HS. We tested lux pR-HS under the same conditions. We can see that the lux pR-HS has similar leakage to Mutant 10, while it has much higher expression intensity. (Figure.3) The statistics of relative induction and leakage intensity of the 10 Mutants and lux pR-HS are shown in Table.1
-
As for why some mutations on -35 to -37 sites would decrease the leakage of lux pR without severely influencing the expression level, we have some hypotheses and explanations. It is reported that -35 to -37 sites are last three bases of lux box which is upstream of the lux promotor and bound specifically by luxR, one of regulatory protein involved in lux expression system.[1] G-36T mutation has less influence than other mutation on sites -35 to -37, which is same to our result. Besides, G-36T is not involved in the two regions of nucleotides -52 to -50 and -39 to -37 which directly contacted with luxR protein. Therefore, this single substitution of nucleotide would decrease the leakage of lux promotor without influencing the expression level too much.[2]
III Protocol
- one Transform the plasmids into E. coli DH5α.
- two Pick a single colony by a sterile tip from each of the LB plates for all the experimental and control groups. Add the colony into 5ml LB medium with ampicillin at 100 ng/µl. Incubate for 6-8 h at 37℃ in a shaker.
- three Measure OD600 of the culture medium with photometer. Dilute the culture medium until OD600 reaches 0.6.
- four Add 100 µl bacteria culture medium into a sterile 96-well plate. Add AHL to final concentrations of 0, 10^-10,10^-9,10^-8 M. Fresh LB medium serves as blank control. Positive control is colony constantly expressing sfGFP and negative control is colony without sfGFP expression. Place the 96-well plate into an automatic microplate reader. Incubate at 16℃ overnight and measure the fluorometric value at 510 nm and OD600 of each well every 30 minutes.
- five Each group should be repeated for at least 3 times.
IV Reference
[1] Antunes, L. C., et al. "A mutational analysis defines Vibrio fischeri LuxR binding sites." Journal of Bacteriology 190.13(2008):4392-4397.
[2] Zeng, Weiqian, et al. "Rational design of an ultrasensitive quorum-sensing switch." Acs Synthetic Biology 6.8(2017).
Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We adapted the part to be able to assemble transcriptional units with the Golden Gate assembly method
Documentation: In order to create our complete [http://2018.igem.org/Team:Valencia_UPV/Part_Collection part collection] of parts compatible with the Golden Gate assembly method, we made the part BBa_K2656003 which is this part adapted to the Golden Gate technology.Results
Sequence and Features
Assembly Compatibility:- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
>Internal Priming Screening Characterization of BBa_R0062: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
=HZAU 2019's characterization=
It is observed that high expression intensity is accompanied by high level of leakage in lux pR promotors. Wild type lux pR represented with black strand in the figures has the highest level of both leakage and intensity. The next three are Mutant 5 (G-36T), Mutant 6 (G-36C), Mutant 9 (T-35G). Mutant 5 has almost the same intensity as wild type, but the leakage is significantly smaller. Mutant 6 and Mutant 9 have a little bit less leakage and medium intensity. We also did the experiment with 10^-9 M and 10^-8 M AHL, all of the promotors show high expression levels and therefore unable to compare. (Data not shown)
//direction/forward
//function/cellsignalling/LuxR
//promoter
//regulation/positive
//rnap/prokaryote/ecoli/sigma70
| control | luxR, HSL |
| direction | Forward |
| negative_regulators | |
| o_h | |
| o_l | |
| positive_regulators | 1 |

 1 Registry Star
1 Registry Star