Part:BBa_K1689001
Coding sequence of STV-N-luc
Design
2015 Peking iGEM combines the homotetrameric streptavidin (STV) protein with the N terminal 416 amino acids of firefly luciferase through a GGGGSGGGGS linker. Biotin-STV interaction is one of the strongest noncovalent interactions in the nature, and is frequently applied as a simple approach to generate semisynthetic DNA-protein conjugates. [1] Split luciferase is particularly useful as split bioluminescent reporter for studying protein-protein interaction. [2] In our dual molecular beacons (MB) detecting system, the fusion protein was incubated with biotinylated MB. And when two parts of split luciferase get closed to each other, they will catalyze the luciferin oxidation to produce luminescent signal that could be detected by microplate reader.
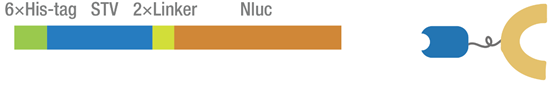
Figure 1 The map of STV-NFluc416 fusion protein.
Expression and purification
This part was constructed on pET28a backbone, placed under T7 promotor and lac operator. Then the recombinant plasmid was transformed into E.coli BL21 strain. However, during protein expression and purification we found that most of the fusion proteins are insoluble in the cell. The inclusion bodies were isolated and then dissolved in 6 mol/L granidine hydrochloride, following by refolding via dilution into pH 8.0 Tris buffer. [3] SDS-PAGE showed the high expression level and high purity of protein.
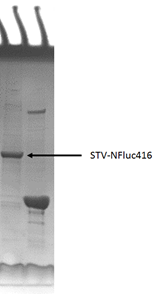
Figure 2 SDS-PAGE of the purified STV-NFluc416 fusion protein.
Characterization
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 4
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |
