Part:BBa_E0033
LacZ alpha fragment; complements matching N-terminal deletion mutant (lacZ-omega)
To greatly reduce the number of bases, this is only a portion of the LacZ gene. It can be used as a detection system with N-terminal deletion mutants of lacZ. Combination of lacZ-alpha with such a deletion mutant protein restores enzyme activity that can be assayed colorimetrically or more sensitively with chemiluminescent substrates.
Improvement by Team:AFCM-EGYPT2022
Improvement by KP-SP tagging
This year, we enhanced the LacZ alpha gene by incorporating a peptide signal that controls the secretion of B galactosidase extracellularly in an effort to improve the cleavage capacity of the X gal, heighten the intensity of the dark blue color emitted by the X gal product, and reduce the amount of time required to receive the results after performing the test on the chip
Improvement of LacZ alpha gene which encodes for the alpha domain of B-galactosidase that acts as a reporter protein in our therapeutic circuit :
Lactose converting enzyme (B-gal) beta-galactosidase that is originated from bacteria And have been studied as a reporter protein for many years ,It's a polypeptide of 1029 amino acid, and its tetramerization carries out the functional form of the enzyme.
The deletion of the tetramerization domain which is the N-terminal region spanning the first 50 residues will abolish its enzymatic activity, which could be fully restored by the alpha complementation of the intact N- terminal portion of the (Beta-gal) this alpha complementation is detected by using a beta-galactosidase colorless substrate as X gal to be cleaved into a dark blue product.
This year we employed this approach to indicate the presence of phenylalanine within our whole cell biosensor on the test line with media containing X gal substrate of our platform to diagnose phenylketonuria
For More Information Check BBa_K4140008
Improvement by Directed Evolution
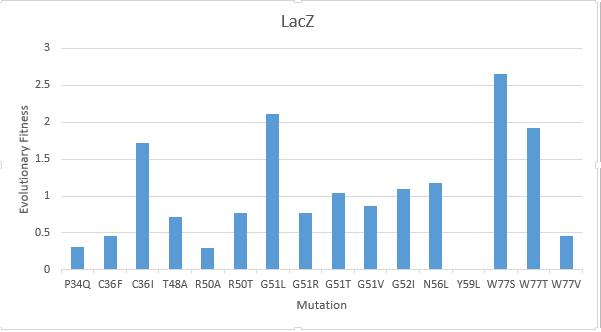
After creating a multiple sequence alignment of the protein sequence and predicting mutational landscapes, the effect of these mutations on the evolutionary fitness of the protein is tested. The prediction of the mutational landscape by saturation mutagenesis of the LacZ protein. The (W77S ) mutation, as depicted in the chart, had the greatest score when compared to other mutations. On the other hand, it's clear that the (Y59L) had the least evolutionary fitness for LacZ protein. As displayed in Figure(2)
New Application (Submitted by Team Michigan 2016)
Alpha complementation is a flexible tool that can be applied for a wide variety of uses where colorimetric detection is desirable. Our team's project design this year proposes to use the in vitro transcription and translation of the alpha fragment as a reporter for proximity dependent ligation. This design involves a pair of ssDNA probes containing aptamer sequences specific to two sites on a target protein. In addition to containing aptamer sequences, these probes also code for the lacZ alpha fragment, but can only express it after a ligation reaction has occurred to join the probes together. The rate of this ligation reaction would be drastically increased when the target biomarker is present to hold the probes in close proximity to one another; therefore expression of the alpha fragment signals the presence of the target protein. The newly produced alpha fragment then complements with the omega fragment (the protein product of the lacZ delta M15 mutation) to form fully functional beta galactosidase which can cleave X-gal to form a blue colorimetric output.


For more information on this application see http://2016.igem.org/Team:Michigan
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 93
Illegal XhoI site found at 144 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//function/coliroid
//function/reporter
//function/reporter/color
| abs | varied |
| emission | |
| emit | |
| excitation | |
| excite | |
| lum | varied |
| protein | LacZ-alpha |
| tag | None |

 1 Registry Star
1 Registry Star