Part:BBa_K112808
Enterobacteria phage T4 Lysis Device - no promoter
This is the lysis device using three genes from enterobacteria phage T4: lysozyme, holin, and antiholin. Various promoters can be installed with this device to control the lysis of bacteria. Antiholin has its own constitutive promoter to prevent the formation of holin multimers from basal expression. When the device is off, higher expression of antiholin compared to holin is required for better stability of the device.
Characterization
For more info, visit [http://2008.igem.org/Team:UC_Berkeley/LysisDevice UC Berkeley iGEM08 Wiki!!]
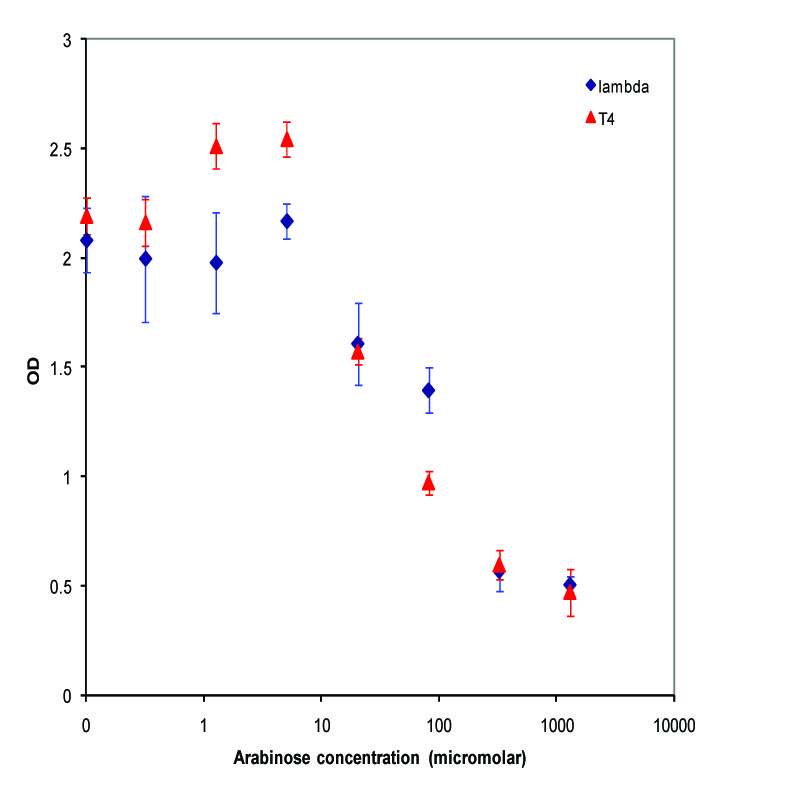
pBAD was assembled with K112022 (Lambda Lysis Device without promoter)and K112808(T4 Lysis Device without promoter), and was inserted into the vector K112950 and pSB1A2, respectively. They were introduced into the MC1061 strain by transformation, and then picked 5 colonies for each device. We grew these cultures to saturation at 37 degrees Celsius in LB media, and then split into eight 1 mL aliquots. A range of concentrations of arabinose was added to these aliquots, with a starting concentration of 1.3E-3 M and the next 6 samples recieving a four-fold dilution of the previous sample, and an equal volume of water added to the last aliquot. The cultures were then incubated at 37 degrees again for 3.5 hours, and the absorbance at 600nm was measured with a Tecan Xfluor4 Safire2 in a Corning Inc. Costar 3603 plate. The data plotted on a log scale is shown below.
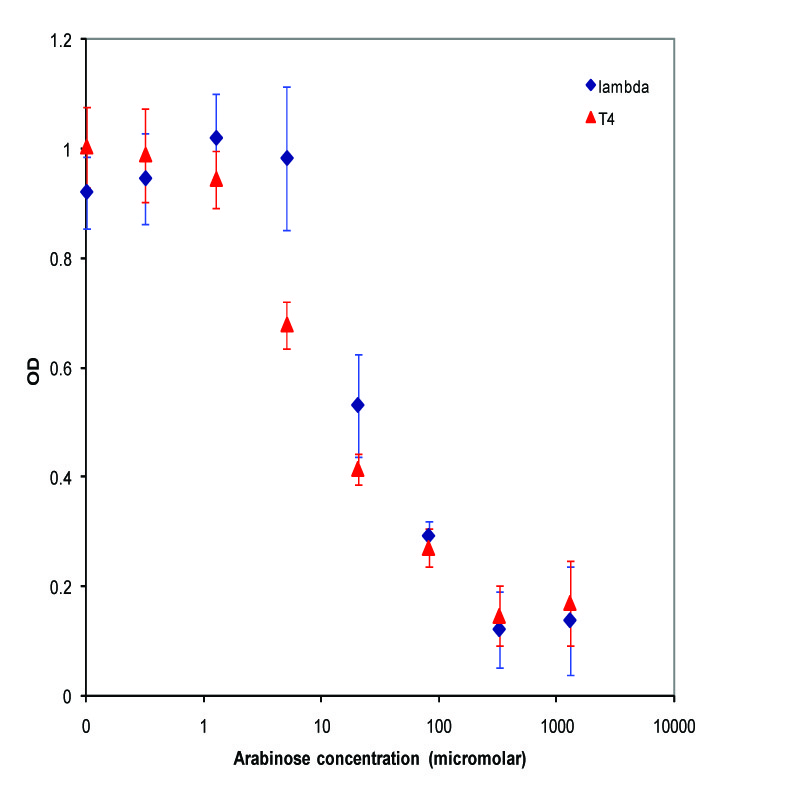
A parallel experiment was performed by taking the same saturated culture before induction with arabinose, diluting 100-fold, growing to mid-log(starting OD .22), before inducing with arabinose as above. The data plotted on a log scale is shown below.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1350
Illegal NheI site found at 1373 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 955
Illegal AgeI site found at 1025 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 1606
| None |

 1 Registry Star
1 Registry Star