Part:BBa_J23106
constitutive promoter family member
Variant RFP (au) J23112 1 J23103 17 J23113 21 J23109 106 J23117 162 J23114 256 J23115 387 J23116 396 J23105 623 J23110 844 J23107 908 J23106 1185 J23108 1303 J23118 1429 J23111 1487 J23101 1791 J23104 1831 J23102 2179 J23100 2547 |
Constitutive promoter family
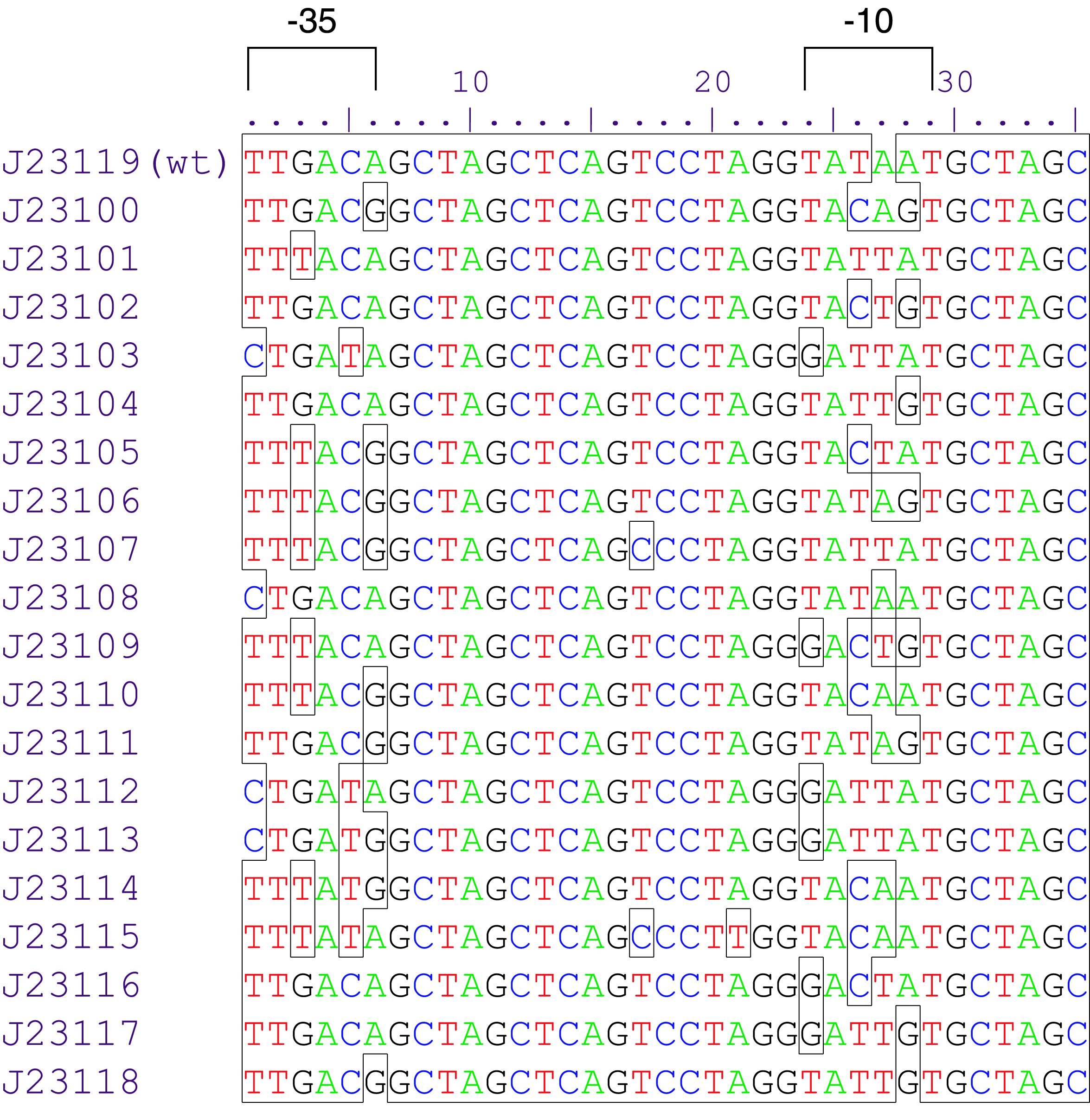
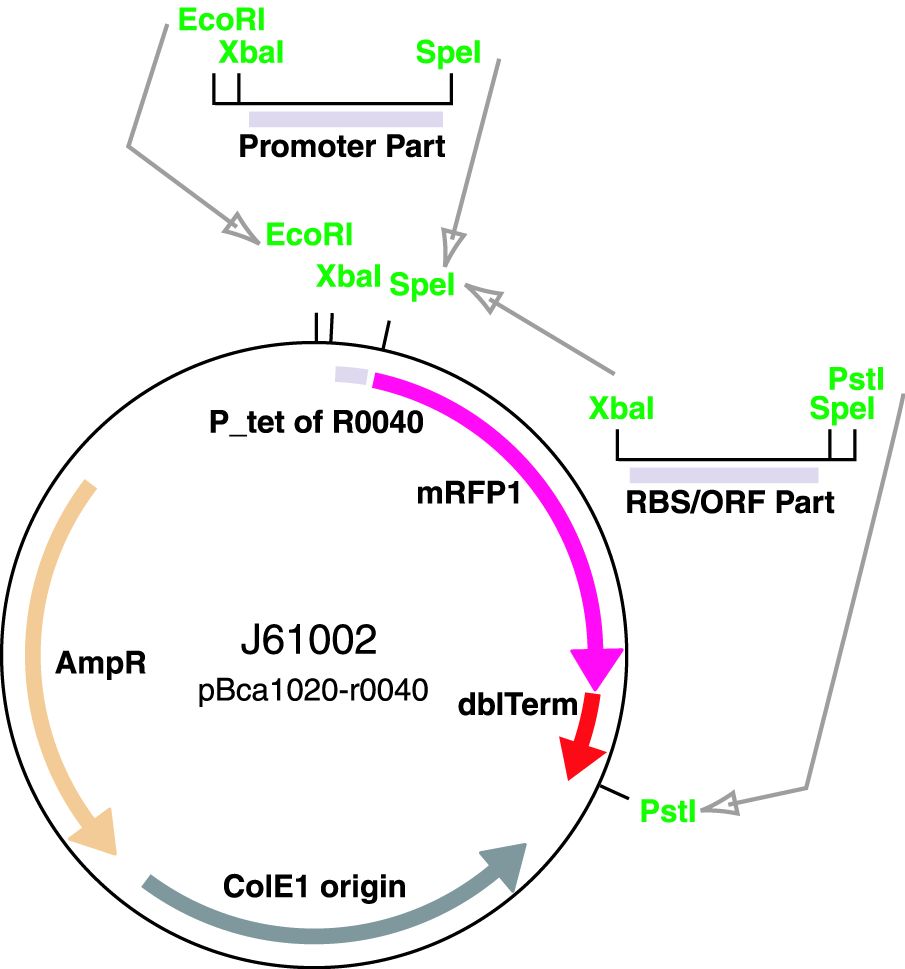
Parts J23100 through J23119 are a family of constitutive promoter parts isolated from a small combinatorial library. J23119 is the "consensus" promoter sequence and the strongest member of the family. All parts except J23119 are present in plasmid J61002. Part J23119 is present in pSB1A2. This places the RFP downstream of the promoter. Reported activities of the promoters are given as the relative fluorescence of these plasmids in strain TG1 grown in LB media to saturation. See part BBa_J61002 for details on their use.
These promoter parts can be used to tune the expression level of constitutively expressed parts. The NheI and AvrII restriction sites present within these promoter parts make them a scaffold for further modification. JCAraw
Jasonk:I suspect that J23102 is in this well rather than J23106, I'd sequence it before using.
Usage and Biology
St Andrews 2018's Characterization
Comparison of the Strength of BBa_J23106 against BBa_J23100
The promoter we assessed was cloned via Gibson Assembly into the pSB1C3 backbone alongside an mNeongreen fluorophore as a reporter. This plasmid was then transformed into DH5alpha super-competent E. coli cells, and the fluorescence and absorbance recorded over the course of 142 hours. The data points were then fitted to a hyperbolic trend line to demonstrate the increase in fluorescence over time. This correlates to the increasing quantity of protein generated, which is directly tied to the strength of the promoter attached.
These studies were done in both LB and M9 media to compare the rate of fluorescence in translucent vs. a transparent medium. As predicted, the M9 medium gave much more reliable results. Below are graphs demonstrating the level of fluorescence generated by the proteins attached to this promoter. It was also compared to the fluorescence generated by a Red Fluorescent Protein (RFP), for some idea of context. A more useful comparison can be made from the graph of concentration, which was derived from the absorbance of each sample. The Red Fluorescent Protein was synthesized using the promoter BBa_J23100, and so the difference in the concentration between the two indicates the relative strengths of the BBa_J23100 and BBa_J23106 promoters. It’s evident that the BBa_J23100 promoter is much stronger than the BBa_J23106 part, producing roughly 13x more RFP than the BBa_J23100 made of the mNeonGreen.
 This graph depicts the difference in fluorescence between the mNeonGreen generated by the BBa_J23106 promoter and the RFP generated by the BBa_J23100 at a wavelength of 650 nm. This study was carried out in transparent M9 media.
This graph depicts the difference in fluorescence between the mNeonGreen generated by the BBa_J23106 promoter and the RFP generated by the BBa_J23100 at a wavelength of 650 nm. This study was carried out in transparent M9 media.
 This graph depicts the difference in fluorescence between the mNeonGreen generated by the BBa_J23106 promoter and the RFP generated by the BBa_J23100 at a wavelength of 515 nm. This study was carried out in transparent M9 media.
This graph depicts the difference in fluorescence between the mNeonGreen generated by the BBa_J23106 promoter and the RFP generated by the BBa_J23100 at a wavelength of 515 nm. This study was carried out in transparent M9 media.
 This graph depicts the difference in fluorescence between the mNeonGreen generated by the BBa_J23106 promoter and the RFP generated by the BBa_J23100 at a wavelength of 650 nm. This study was carried out in brown LB media.
This graph depicts the difference in fluorescence between the mNeonGreen generated by the BBa_J23106 promoter and the RFP generated by the BBa_J23100 at a wavelength of 650 nm. This study was carried out in brown LB media.
 This graph depicts the difference in fluorescence between the mNeonGreen generated by the BBa_J23106 promoter and the RFP generated by the BBa_J23100 at a wavelength of 515 nm. This study was carried out in brown LB media.
This graph depicts the difference in fluorescence between the mNeonGreen generated by the BBa_J23106 promoter and the RFP generated by the BBa_J23100 at a wavelength of 515 nm. This study was carried out in brown LB media.
 This graph shows the concentration of protein derived from the absorbance and the extinction coefficient of each respective fluorophore.
This graph shows the concentration of protein derived from the absorbance and the extinction coefficient of each respective fluorophore.
Sheffield 2016's Characterisation
Measured strength
Sheffield 2016 has improved the characterisation of both BBa_J23100 and BBa_J23106. These parts are a strong and medium promoter respectively, that we have used to design our iron detecting device. We have experimentally validated through fluorimetry that there is indeed a significant difference between expression levels of GFP coupled to the strong and medium promoters. Comparative analysis of promoter strengths can be directly interpreted from the data we obtained. This data can be found both on the original part experience pages of BBa_J23100 and BBa_J23106, as well as on our website.

Fluorescence of JC28 mutants or W3110 wild types transformed with RyhB-GFP constructs under the control of medium (MedGFP) or strong promoters (StrGFP).
Determination of Noise Levels in Constitutive Promoter Family Members
(characterized by SDU-Denmark 2017)
Fluorescence microscopy and flow cytometry revealed decrease in fluorescence over time for members of the constitutive promoter family.
The expression levels and the noise of four different members of the Anderson promoter collection and their RFP reporter systems, were studied by fluorescence microscopy. These were, in increasing promoter strength, BBa_J23114, BBa_J23110, BBa_J23106, and BBa_J23102
Additionally, the change in RFP expression levels and noise during growth were tested for the promoters with the highest and lowest relative promoter strength by flow cytometry and qualitative analysis by fluorescence microscopy. Combining these two techniques, the expression and noise levels for the promoters were determined as follows:
- The weak promoter, BBa_J23114, exhibited a relatively low expression of RFP, indicating low gene expression and an increasing high level of noise throughout growth.
- Both medium strength promoters, BBa_J23110 and BBa_J23106, displayed a moderate level of both noise and protein expression of the RFP reporter.
- The strong promoter, BBa_J23102, exhibited a comparatively high expression of the reporter RFP and an increasing high level of noise throughout growth.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
Relative promoter strength estimates (see [http://2009.igem.org/Team:Groningen/Promoters this page] from Groningen 2009):
| Reference | Strength |
|---|---|
| BBa_J23100 | 0.49 |
| BBa_J23109 | 33 |
Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We adapted the part to be able to assemble transcriptional units with the Golden Gate assembly method
Documentation:
In order to create our complete [http://2018.igem.org/Team:Valencia_UPV/Part_Collection part collection] of parts compatible with the Golden Gate assembly method, we made the part BBa_K2656004 which is this part adapted to the Golden Gate technology.
>Internal Priming Screening Characterization of BBa_J23106: Has no possible internal priming sites between this BioBrick part and the VF2 or the VR primer.
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
Thessaly 2019 Characterization
Thessaly 2019 sought to characterize the coding sequence of TEM-optimized beta-lactamase (BBa_I757010) under the regulation of the constituve Anderson Family promoters BBa_J23100, BBa_J23105, BBa_J23106, BBa_J23119. Beta-lactamase is an enzyme that hydrolyses beta-lactams (e.g. ampicillin) and is naturally found in procaryotic cells. A colorimetric assay has been developed using nitrocefin as a substrate which after hydrolysis from beta-lactamase changes the reaction color, from yellow (380nm) to red (490nm).
To achieve that, the coding sequence was assembled with each promoter, a universal RBS (BBa_B0034) and a double terminator(BBa_B0015). The parts were cloned in pSB1C3 and pSB1K3 and transformed into E. coli DH5a competent cells. For protein expression, the plasmids were transformed into E. coli BL21 (DE3) competent cells.
For the beta-lactamase assay, we set up the following experimental design:
1. Grow BL21 (DE3) cells overnight in 5ml LB (~16h) at a shaken incubator, 37 degrees C / 210rpm
2. The following morning, measure the OD600 of overnight cultures
3. Dilute all cultures to OD600¬ = 0.05 in M9 minimal medium
4. Grow cells 37 degrees C /210 RPM until OD600=0.4-0.6 (~2h)
5. Dilute all cells to the same OD600 (e.g. 0.4)
6. Load 160 of culture in a 96-well plate (do triplicates). Add 40 ul 0.5 uM nitrocefin for a final concentration of 100nM
7. Measure the absorbance at 490nm (for nitrocefin hydrolysis) and 600nm (for cell growth) every 30 seconds for 25 minutes in a microplate reader. Shake between measurements.
To ensure that the absorbance shown corresponds only to enzymatic activity by beta-lactamase, we included 3 controls in the experiment. The first control has M9 medium only (no cells) and nitrocefin, the second has empty BL21 (DE3) cells (no plasmid) and nitrocefin, while the third has BL21 (DE3) cells containing the plasmid but not the part (empty plasmid). To obtain comparable results, we normalized all values by dividing OD490 by OD600.
The results are shown in the graph below

XJTU-CHINA 2019 Characterization
XJTU-CHINA 2019 uses pJ23106 as a constitutive promoter of a photosensitive protein -- Cph8 (BBa_K2598006) which is the central part of our light-control system, so we plan to quantitatively characterize this promoter BBa_J23106 in order to acquire parameters in realistic conditions which can be used in modelling. We use part BBa_K1819006, which is consist of GFP (BBa_K1819000) and its promoter J23106. We have experimentally validated this part in three different chassis which are three strains of E.coli: DH5 α, TG1 and BL21.
For the J23106 strength assay, we set up the following experimental design:
Day1: Transform Escherichia coli DH5 α, TG1 and BL21 with pSB1K3-K1819006.
Day2: Pick colony from each of the transformation plates and inoculate in 5-10 mL LB medium + Chloramphenicol. Grow the cells for 12h.
Day3:
1. Mix 0.4ml bacterial solution with 20ml LB+20ul Chlo;
2. Culture the diluted bacterial solution at 37℃,210 RPM;
3. Measure OD and FL of the bacterial solution and LB + Chloramphenicol medium every 1 hour (With 3*6 parallel groups and 6 control groups, 200 ul of fluid were placed in each hole, 24 holes in total).
//direction/forward
//promoter/anderson
//regulation/constitutive
//rnap/prokaryote/ecoli/sigma70
| negative_regulators | |
| positive_regulators |

 1 Registry Star
1 Registry Star