Part:BBa_K2065000
CT52-SmallBiT
The SmallBiT-CT52 BioBrick is part of a split luciferase system. This luciferase system consists of two parts, namely SmallBiT and LargeBiT. It is a ATP-independent luciferase, that produces high intensity bioluminescence. The total weight of this luciferase system is 19.1 kDa. The NanoLuc system consists of 171 amino acids. When they dimerize, this means when they come into close proximity of each other, one can see bioluminescence. The emission peak of this NanoBiT system is around 460 nanometers. This is blue light.
Usage and Biology
SmallBiT is linked to CT52 with a GGS10-linker, so it actually consists of three subparts. SmallBiT is 33 basepairs long and thus consists of 11 amino acids. The GGS10-linker that is used, consists of 30 amino acids, it is a ten-times repeated sequence of glycine-glycine-serine amino acids. This complex is linked to CT52. This protein consist of the last 52 amino acids of the C-terminal region of the H+-ATPase. The N-terminus of CT52 is a free end, which can be used to link proteins to. The binding between CT52 and T14-3-3 is stabilized by the small molecule Fusicoccin. When Fusicoccin is present, the affinity of CT52 for T14-3-3 is increased in 30 fold with respect to the situation when Fusicoccin is not present [3]. The binding interaction with Fusicoccin is reversibel, this means CT52 will dissociate from the scaffold.
This means that the CT52 -SmallBiT complex will move towards the scaffold protein, after a small molecule, Fusicoccin, is added. Three different variants of the CT52-protein were created with the help of the Rosetta software package. Each mutation in the CT52 protein has its complementary mutation in the T14-3-3 protein. Because these mutations were found with the help of the Rosetta software package, the quality of these mutations have to be verified. One variant of the CT52 protein was already known and tested. This mutation functioned as a positive control in the verification process of the other mutations.
Sequence
The sequence of our CT52-GGS10-SmallBiT has been verified by StarSeq. It contains the prefix and suffix with the correct restriction sites (EcoRI, XbaI, SpeI and PstI). CT52-GGS10-smallBiT is 279 basepairs long.
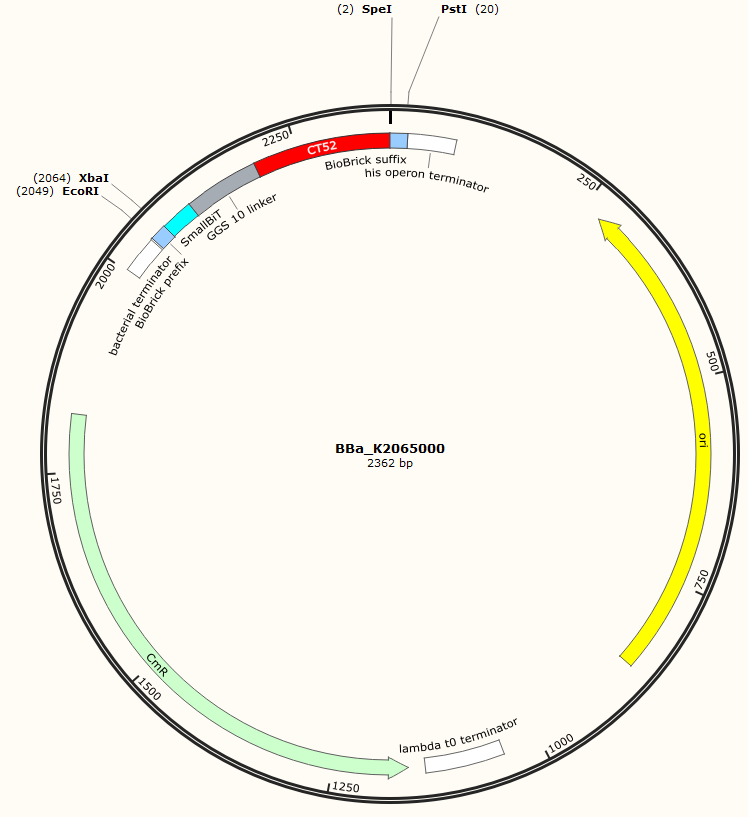
Figure 1: Snapgene map BBa_K2065000
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 289
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
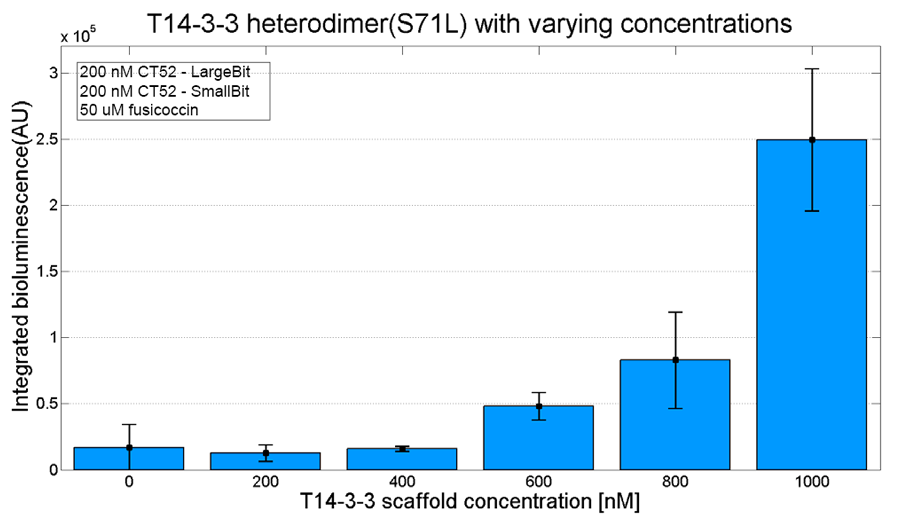
Bioluminescence confirmation
To determine whether the split luciferase system is working, one should see a bioluminescence signal when the two components come into close proximity of each other. A bioluminescence peak around 460nm must be visible when SmallBiT and LargeBiT dimerize.
When the two NanoBiTs are into close proximity of each other, they dimerize. When Furimazine is added to the dimerized Nanoluc luciferase, the Furimazine is converted into Furimamide. When this reaction occurs, light with a wavelength of 460 nm will be emitted.
The spectrum of the CT52-NanoBiT system is visualed below
CT52-SmallBiT Statistics
| Protein Specifications CT52-SmallBiT | |||
|---|---|---|---|
| General Information | Number of amino acids | 93 | |
| Molecular weight | 9367.169 | ||
| Theoretical pi | 5.62 | ||
| Extinction coefficient | 2980 | ||
| Formula | C395H639N123O141 | ||
| Total numbers of atons | 1298 | ||
| Amino Acid Composition | Amino Acid | Frequency | Percentage(%) |
| Ala(A) | 4 | 4.3 | |
| Arg(R | 5 | 5.4 | |
| Asn(N) | 3 | 3.2 | |
| Asp(D) | 2 | 2.2 | |
| Cys(C) | 0 | 0 | |
| Gln(Q) | 4 | 4.3 | |
| Glu(E) | 9 | 9.7 | |
| Gly(G) | 23 | 24.7 | |
| His(H) | 2 | 2.2 | |
| Ile(I) | 5 | 5.4 | |
| Leu(L) | 8 | 8.6 | |
| Lys(K) | 4 | 4.3 | |
| Met(M) | 0 | 0 | |
| Phe(F) | 2 | 2.2 | |
| Pro(P) | 0 | 0 | |
| Ser(S) | 12 | 12.9 | |
| Thr(T) | 4 | 4.3 | |
| Trp(W) | 0 | 0 | |
| Tyr(T) | 2 | 2.2 | |
| Val(V) | 4 | 4.3 | |
| Pyl(O) | 0 | 0.0 | |
| Sec(U) | 0 | 0.0 | |
| None |
