Part:BBa_K592012
eforRed, red chromoprotein
This chromoprotein eforRed naturally exhibits red/pink color when expressed. The color is slightly weaker than RFP. On agar plates and in liquid culture, the color is readily visible to naked eye in less than 24 hours of incubation. The DNA was codon-optimized for expression in E.coli and synthesized by the Korean company Bioneer Corporation.
This part can also be found as a coding device BBa_K864402.
Usage and Biology
This part is useful as a reporter.
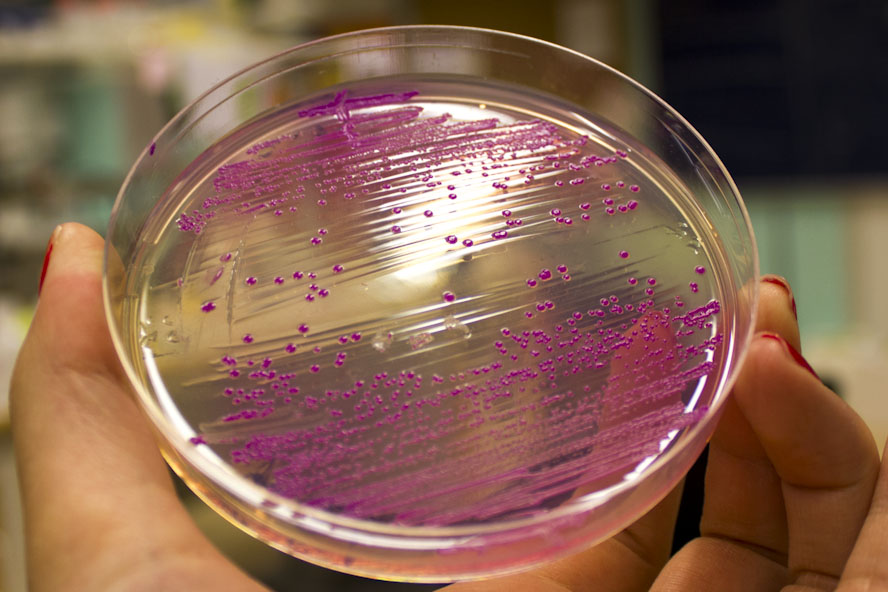
 iGEM12_Uppsala_University: Left picture: Visual color expression of eforRed (BBa_K592012).
iGEM12_Uppsala_University: Left picture: Visual color expression of eforRed (BBa_K592012).
Right picture: The Uppsala chromoprotein collection and RFP. The image shows pellets of E coli expressing chromoproteins eforRed BBa_K592012, RFP BBa_E1010, cjBlue BBa_K592011, aeBlue BBa_K864401, amilGFP BBa_K592010 and amilCP BBa_K592009.
Getting this part
All chromoproteins displayed above should be available, but if the registry cannot send them, feel free to contact the Uppsala University team directly via erik.gullberg(at)imbim.uu.se, and we will send them to you.
References
[http://www.ncbi.nlm.nih.gov/pubmed/18648549] Alieva, N. O., et al. 2008. Diversity and evolution of coral fluorescent proteins. PLoS One 3:e2680.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| Protein data table for BioBrick BBa_K592012 automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCATGTCAGTG ... GCTCTCCCAACCGGT ORF from nucleotide position -8 to 687 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| Alignments (obtained from PredictProtein.org) There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
| There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
//collections/chromoprotein/uppsala
//function/reporter/pigment
| None |
