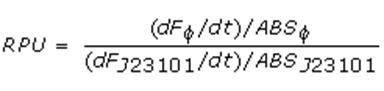Difference between revisions of "Part:BBa R0011:Experience"
(→User Reviews) |
(→User Reviews) |
||
| Line 194: | Line 194: | ||
RFP generator (<partinfo>BBa_I13507</partinfo>) was used as a reporter gene. In particular, these measurement systems were used: | RFP generator (<partinfo>BBa_I13507</partinfo>) was used as a reporter gene. In particular, these measurement systems were used: | ||
| − | <partinfo>pSB1A2</partinfo>-<partinfo>BBa_J107010</partinfo> | + | |
| − | <partinfo>pSB1A2</partinfo>-<partinfo>BBa_J04450</partinfo> | + | *<partinfo>pSB1A2</partinfo>-<partinfo>BBa_J107010</partinfo> |
| − | <partinfo>pSB4C5</partinfo>-<partinfo>BBa_J107010</partinfo> | + | *<partinfo>pSB1A2</partinfo>-<partinfo>BBa_J04450</partinfo> |
| − | <partinfo>pSB4C5</partinfo>-<partinfo>BBa_J04450</partinfo> | + | *<partinfo>pSB4C5</partinfo>-<partinfo>BBa_J107010</partinfo> |
| + | *<partinfo>pSB4C5</partinfo>-<partinfo>BBa_J04450</partinfo> | ||
At first, <partinfo>BBa_J107010</partinfo> and <partinfo>BBa_J04450</partinfo> inducibility was tested in a high copy number vector (<partinfo>pSB1A2</partinfo>). The results are shown here as the relative RFP synthesis rate per cell. | At first, <partinfo>BBa_J107010</partinfo> and <partinfo>BBa_J04450</partinfo> inducibility was tested in a high copy number vector (<partinfo>pSB1A2</partinfo>). The results are shown here as the relative RFP synthesis rate per cell. | ||
| Line 205: | Line 206: | ||
| − | Results show that in this condition <partinfo>BBa_R0010</partinfo> is about 2 fold stronger than <partinfo>BBa_R0011</partinfo>, but induced and uninduced cultures did not show differences in the RFP signal. | + | Results show that in this condition <partinfo>BBa_R0010</partinfo> is about 2-fold stronger than <partinfo>BBa_R0011</partinfo>, but induced and uninduced cultures did not show differences in the RFP signal. |
This result is expected because the <partinfo>pSB1A2</partinfo> is propagated at about 200 copies per cell, while the lacI repressor is present at single copy in the genome and thus it is not able to repress the lac promoters. | This result is expected because the <partinfo>pSB1A2</partinfo> is propagated at about 200 copies per cell, while the lacI repressor is present at single copy in the genome and thus it is not able to repress the lac promoters. | ||
| Line 218: | Line 219: | ||
Data show that <partinfo>BBa_R0011</partinfo> is stronger than the <partinfo>BBa_R0010</partinfo> wild type promoter in low copy plasmid. In the uninduced state, <partinfo>BBa_R0011</partinfo> has about the same strength as the <partinfo>BBa_J23101</partinfo> reference standard promoter. | Data show that <partinfo>BBa_R0011</partinfo> is stronger than the <partinfo>BBa_R0010</partinfo> wild type promoter in low copy plasmid. In the uninduced state, <partinfo>BBa_R0011</partinfo> has about the same strength as the <partinfo>BBa_J23101</partinfo> reference standard promoter. | ||
This static characteristic shows that the promoters are both leaky and a very low IPTG concentration (10 uM) is sufficient to trigger gene expression at *very* high levels. | This static characteristic shows that the promoters are both leaky and a very low IPTG concentration (10 uM) is sufficient to trigger gene expression at *very* high levels. | ||
| + | |||
| + | These results demonstrate that the genomic lacI is partially able to repress the two promoters, but very low IPTG concentrations are sufficient to bind the repressor and trigger the promoters transcription. | ||
| + | |||
| + | |||
| + | '''Dynamic characterization in low copy vector:''' | ||
| + | |||
| + | FIGURA | ||
| + | |||
'''Conclusion:''' the characterization of two IPTG-inducible promoters has been performed and the performance of these two promoters have been compared in terms of transcriptional strength. The reported results are easily sharable in different laboratories thanks to the used standard RPU approach. | '''Conclusion:''' the characterization of two IPTG-inducible promoters has been performed and the performance of these two promoters have been compared in terms of transcriptional strength. The reported results are easily sharable in different laboratories thanks to the used standard RPU approach. | ||
Revision as of 22:18, 26 October 2010
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_R0011
BBa_R0011 - Plac hybrid promoter - UNIPV-Pavia team (test performed by L. Pasotti, S. Zucca)
Description
Here we provide the characterization of this promoter in E. coli TOP10, which has a lacI genomic copy, constitutively expressed in a weak manner.
The data below are referred to BBa_K173025, which is the measurement system of BBa_R0011.
Characterization
Compatibility: E. coli TOP10 in pSB1A2.
| IPTG concentration |
LB | |
| Doubling time [minutes] | RPU | |
| 0uM | 42 | 1.52 [1.46; 1.59] |
| 10uM | 41 | 1.83 [1.73; 1.90] |
| 50uM | 43 | 1.78 [1.64 ; 1.92] |
| 100uM | 46 | 1.73 [1.52; 1.95] |
| 500uM | 43 | 1.61 [1.46 ; 1.81] |
| 1mM | 47 | 1.78 [1.53; 2.01] |
| 2mM | 44 | 1.47 [0.93; 2.05] |
 BBa_K173025 Growth curves for BBa_K173025 in LB |
 BBa_K173025(dGFP/dt)/O.D. in LB |
 BBa_K173025 Induction curve of BBa_K173025 in LB |
Conclusions
As TOP10 strains contains an expressed lacI in its genome, we wanted to know if it was enough to repress Plac activity in a high copy number plasmid in order to use it as a lactose/IPTG sensor. It is evident from induction curve that the behaviour of this part is almost independent from IPTG concentration, probably because lacI production by genomic DNA is too low. So our characterization has shown that BBa_R0011 can be used as a strong constitutive promoter, with mean RPU higher than 1.5. Future work should be dedicated to the characterization of this BioBrick in low copy number plasmids and in strains containing lacIq mutation in order to buil-up new IPTG/lactose sensors.
Growth conditions
Microplate reader experiments
- 8 ul of long term storage glycerol stock were inoculated in 5 ml of LB + suitable antibiotic in a 15 ml falcon tube and incubated at 37°C, 220 rpm for about 16 hours.
- The grown cultures were then diluted 1:100 in 5 ml of LB or M9 supplemented medium and incubated in the same conditions as before for about 4 hours.
- These new cultures were diluted to an O.D.600 of 0.02 (measured with a TECAN F200 microplate reader on a 200 ul of volume per well; it is not comparable with the 1 cm pathlength cuvette) in a sufficient amount of medium to fill all the desired microplate wells.
- These new dilutions were aliquoted in a flat-bottom 96-well microplate, avoiding to perform dynamic experiments in the microplate frame (see [http://2009.igem.org/Team:UNIPV-Pavia/Methods_Materials/Evaporation Frame effect section] for details). All the wells were filled with a 200 ul volume.
- If required, 2 ul of inducer were added to each single well.
- The microplate was incubated in the Tecan Infinite F200 microplate reader and fluorescence (when required) and absorbance were measured with this automatic protocol:
- 37°C constant for all the experiment;
- sampling time of 5 minutes;
- fluorescence gain of 50;
- O.D. filter was 600 nm;
- GFP filters were 485nm (ex) / 540nm (em);
- 15 seconds of linear shaking (3mm amplitude) followed by 10 seconds of waiting before the measurements in order to make a homogeneous culture.
- Variable experiment duration time (from 3 to 24 hours).
Data analysis
Growth curves
All our growth curves have been obtained subtracting for each time sample the broth O.D.600 measurement from that of the culture; broth was considered in the same conditions of the culture (e.g. induced with the same inducer concentration).
Doubling time
The natural logarithm of the growth curves (processed according to the above section) was computed and the linear phase (corresponding to the bacterial exponential growth phase) was isolated by visual inspection. Then the linear regression was performed in order to estimate the slope of the line m. Finally the doubling time was estimated as d=ln(2)/m [minutes].
In the case of multiple growth curves for a strain, the mean value of the processed curves was computed for each time sample before applying the above described procedure.
Relative Promoter Units (RPUs)
The RPUs are standard units proposed by Kelly J. et al., 2008, in which the transcriptional strength of a promoter can be measured using a reference standard, just like the ground in electric circuits.
RPUs have been computed as:
in which:
- phi is the considered promoter and J23101 is the reference standard promoter (taken from Anderson Promoter Collection);
- F is the blanked fluorescence of the culture, computed subtracting for each time sample fluorescence measure for negative control from that of culture, where the negative control is a non-fluorescent strain (in our experiment it is usually used TOP10 strain bearing BBa_B0032 or BBa_B0033, which are symmply RBSs do not have expression systems for reporter genes);
- ABS is the blanked absorbance (O.D.600) of the culture, computed as described in "Growth curves" section.
RPU measurement has the following advantages (under suitable conditions)
- it is proportional to PoPS (Polymerase Per Second), a very important parameter that expresses the transcription rate of a promoter;
- it uses a reference standard and so measurements can be compared between different laboratories.
The hypotheses on which RPU theory is based can be found in Kelly J. et al., 2008, as well as all the mathematical steps. From our point of view, the main hypotheses that have to be satisfied are the following:
- the reporter protein must have a half life higher than the experiment duration (we use GFPmut3, BBa_E0240, which has an estimated half life of at least 24 hours, and the experiments duration is always less than 7 hours);
- strain, plasmid copy number, antibiotic, growth medium, growth conditions, protein generator assembled downstream of the promoter must be the same in the promoter of interest and in J23101 reference standard.
- steady state must be valid, so (dF/dt)/ABS (proportional to the GFP synthesis rate per cell) must be constant.
Inducible systems
Every experiment is performed on the following cultures:
- the culture of interest (system studied expressing GFP)
- the benchmarck used to evaluate R.P.U. (BBa_K173001 measurement part, that is BBa_J23101 with BBa_E0240 downstream)
- a negative control (generally, BBa_B0033 RBS)
For inducible systems several plots are reported. The first plot is a panel containing 4 subplots, numerated this way:
| (1) | (2) |
| (3) |
Plot (1) contains growth curves of the cultures, after blank value has been removed. Every curve is calculated averaging on three replicates of the same culture and subtracting the blank for each time sample. Blank is calculated averaging the replicates of blank wells.
Plot (2) shows the logarithm of absorbance in exponential phase of bacterial growth, determined by a visual inspection of log-plots. These values are used to evaluate doubling time and R.P.U..
Plot (3) contains (dGFP/dt)/O.D., the value named S_cell in Kelly J. et al., 2008 procedure for RPU evaluation.
In these plots are reported black veritcal lines that define the range of values used to evaluate RPU. It is important to underline, as explained in next paragraph, that RPU are calculated on cultures at the same O.D. level, not at the same time.
The second graphic shows S_cell VS O.D.. This plot allows the conparison of S_cell values between different cultures, that are supposed to reach the same level of growth not at the same time, but at the same O.D. value.
The third graphic shows the induction curve. The RPU value is calculated on S_cell values corresponding to O.D. values in exponential phase (typically, from 0.05 to 0.16). The curve is obtained averaging in time S_cell values corresponding to exponential phase.
Error bars rapresent the minimum and maximum value of R.P.U. belonging to the range of O.D. in exponential phase.
Materials
- Long term glycerol stocks were stored at -80°C with a final glycerol concentration of 20%
- Antibiotics were: Ampicillin (Amp) 100 ug/ml stored at -20°C in 1000x stocks. Amp was dissolved in water.
- LB medium was prepare with: 1% NaCl, 1% bactotryptone, 0.5% yeast extract. The medium was not buffered with NaOH.
- M9 supplemented medium was prepared according to: [http://openwetware.org/wiki/Knight:M9_supplemented_media Openwetware protocol].
- Ready made IPTG (Sigma) was stored at -20°C in a 200mM stock.
User Reviews
UNIQ346d0fda2e7c35f7-partinfo-0000000E-QINU
|
•••••
Antiquity |
This review comes from the old result system and indicates that this part worked in some test. |
|
•••
UNIPV-Pavia iGEM 2010 |
BBa_R0011 hybrid lac promoter and the BBa_R0010 wild type lac promoter were characterized at different copy number in TOP10 E. coli strain. This strain contains a lacI expression system in the genome. Induction static transfer function (computed in Relative Promoter Units), dynamics and metabolic burden were evaluated as a function of different IPTG concentrations in M9 supplemented with glycerol growth medium. RFP generator (BBa_I13507) was used as a reporter gene. In particular, these measurement systems were used: At first, BBa_J107010 and BBa_J04450 inducibility was tested in a high copy number vector (pSB1A2). The results are shown here as the relative RFP synthesis rate per cell.
This result is expected because the pSB1A2 is propagated at about 200 copies per cell, while the lacI repressor is present at single copy in the genome and thus it is not able to repress the lac promoters.
FIGURA Results show that in this condition both BBa_R0010 and BBa_R0011 produce different amounts of RFP as a function of the IPTG concentration. The amplitude of the two curves show that the promoters are very strong when induced with IPTG >= 10 uM. Data show that BBa_R0011 is stronger than the BBa_R0010 wild type promoter in low copy plasmid. In the uninduced state, BBa_R0011 has about the same strength as the BBa_J23101 reference standard promoter. This static characteristic shows that the promoters are both leaky and a very low IPTG concentration (10 uM) is sufficient to trigger gene expression at *very* high levels. These results demonstrate that the genomic lacI is partially able to repress the two promoters, but very low IPTG concentrations are sufficient to bind the repressor and trigger the promoters transcription.
FIGURA
|
UNIQ346d0fda2e7c35f7-partinfo-0000002B-QINU