Difference between revisions of "Part:BBa K346002"
| Line 17: | Line 17: | ||
The key sequence for MerR’s binding is a region of interrupted dyad symmetry (19bp) located between the -35 and -10 haxamers of Ptpcad (The top strand). And the structure of Pr (botton strand) is similar to Ptpcad in a divergent orientation. The -10 hexamers of Ptpcad and Pr actually overlap by four bases. When the apo-MerR dimer bind to the dyad symmetrical operator DNA between the -35 and – 10 elements of mercury inducible promoter, PmerT, which has a unusually long spacer of 19 bp for MerR to bind on, the binding of RNA polymerase is inhibited(Fig.2). When Hg(II) is available in the environment, the ion binds to merR between the two subunits. The Hg-bound MerR can result in an a structural distortion of PmerT, allowing the RNA polymerase contacts to be made, leading to the expression of down-stream genes. This mode of transcription activation indicates that as the apo-MerR and Hg-bound MerR have a competing relationship. The initiation of PmerT is controlled by the expression level of MerR, and the activity of which can be denoted by reporter genes. Thus the sensitivity of Hg(II) in cell can be manipulated. | The key sequence for MerR’s binding is a region of interrupted dyad symmetry (19bp) located between the -35 and -10 haxamers of Ptpcad (The top strand). And the structure of Pr (botton strand) is similar to Ptpcad in a divergent orientation. The -10 hexamers of Ptpcad and Pr actually overlap by four bases. When the apo-MerR dimer bind to the dyad symmetrical operator DNA between the -35 and – 10 elements of mercury inducible promoter, PmerT, which has a unusually long spacer of 19 bp for MerR to bind on, the binding of RNA polymerase is inhibited(Fig.2). When Hg(II) is available in the environment, the ion binds to merR between the two subunits. The Hg-bound MerR can result in an a structural distortion of PmerT, allowing the RNA polymerase contacts to be made, leading to the expression of down-stream genes. This mode of transcription activation indicates that as the apo-MerR and Hg-bound MerR have a competing relationship. The initiation of PmerT is controlled by the expression level of MerR, and the activity of which can be denoted by reporter genes. Thus the sensitivity of Hg(II) in cell can be manipulated. | ||
| − | [[Image: | + | [[Image:PmerT2.jpg]] |
'''Fig.2. DNA sequence of the Tn21 mer operator promoter region.''' The MerR binding site on PmerT is marked by a box. The -35 and -10 regions for both PmerR and PmerTPAD are marked with boxes, and the dyad symmetrical DNA sequence that MerR recognizes and binds to is marked with arrows under the DNA sequence. The divergently oriented promoters are marked by blue box and purple box, respectively. | '''Fig.2. DNA sequence of the Tn21 mer operator promoter region.''' The MerR binding site on PmerT is marked by a box. The -35 and -10 regions for both PmerR and PmerTPAD are marked with boxes, and the dyad symmetrical DNA sequence that MerR recognizes and binds to is marked with arrows under the DNA sequence. The divergently oriented promoters are marked by blue box and purple box, respectively. | ||
Revision as of 18:20, 26 October 2010
PmerT promoter (mercury-responsive)
This part, PmerT, is a promoter from Tn21 mercury resistance (mer) operon. The mer operon of Tn21 consists of two tightly overlapped, divergently oriented promoters – Pr and Ptpad.(Park, Wireman et al. 1992). Pr is the promoter of the regulatory protein gene, merR, and Ptpcad is for the transcription of the structural gene – merPTAD. They are called merOP as a whole.
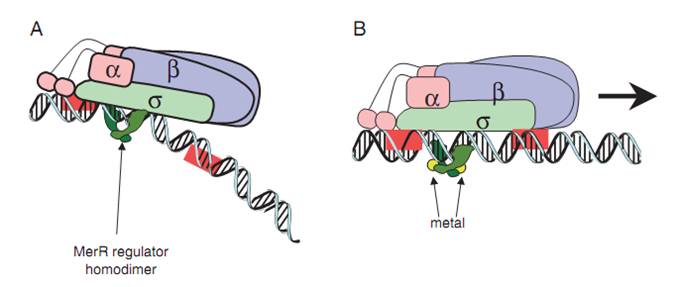
merOP is bound by MerR, a regulatary protein, as a homodimer and the occupancy of Ptpad by RNA polymerase is enhanced regardless of the presence of Hg(II), although only after Hg(II)’s binding can the dimer stop preventing the formation of the open complex by RNA polymerase(Fig.1).
Fig.1. A generalized mechanism of MerR family regulator transcriptional activation. (A) The dimeric MerR regulator binds to the operator region of the promoter and recruits RNA polymerase, forming a ternary complex. Transcription is slightly repressed because the apo-MerR regulator dimer has bent the promoter DNA such that RNA polymerase does not contact it properly. (B) Upon binding the cognate metal ions (shown as cyan circles) the metallated MerR homodimer causes a realignment of the promoter such that RNA polymerase contacts the -35 and -10 sequences leading to open complex formation and transcription. Modified from Brown et al.
The key sequence for MerR’s binding is a region of interrupted dyad symmetry (19bp) located between the -35 and -10 haxamers of Ptpcad (The top strand). And the structure of Pr (botton strand) is similar to Ptpcad in a divergent orientation. The -10 hexamers of Ptpcad and Pr actually overlap by four bases. When the apo-MerR dimer bind to the dyad symmetrical operator DNA between the -35 and – 10 elements of mercury inducible promoter, PmerT, which has a unusually long spacer of 19 bp for MerR to bind on, the binding of RNA polymerase is inhibited(Fig.2). When Hg(II) is available in the environment, the ion binds to merR between the two subunits. The Hg-bound MerR can result in an a structural distortion of PmerT, allowing the RNA polymerase contacts to be made, leading to the expression of down-stream genes. This mode of transcription activation indicates that as the apo-MerR and Hg-bound MerR have a competing relationship. The initiation of PmerT is controlled by the expression level of MerR, and the activity of which can be denoted by reporter genes. Thus the sensitivity of Hg(II) in cell can be manipulated.
Fig.2. DNA sequence of the Tn21 mer operator promoter region. The MerR binding site on PmerT is marked by a box. The -35 and -10 regions for both PmerR and PmerTPAD are marked with boxes, and the dyad symmetrical DNA sequence that MerR recognizes and binds to is marked with arrows under the DNA sequence. The divergently oriented promoters are marked by blue box and purple box, respectively.
For more information, please check 'Experience' and our wiki!
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]