Difference between revisions of "Part:BBa K346001"
| Line 8: | Line 8: | ||
The mercury resistance operon, ''mer'', can enable bacteria to avoid and remove toxic metal Hg under the regulation of the MerR family transcriptional factor MerR. MerR acts as an activator of mer genes in response to the presence of Hg (II), while it will be reversed into a weak repressor in the absence of Hg (II), to maintain its own expression at certain level. | The mercury resistance operon, ''mer'', can enable bacteria to avoid and remove toxic metal Hg under the regulation of the MerR family transcriptional factor MerR. MerR acts as an activator of mer genes in response to the presence of Hg (II), while it will be reversed into a weak repressor in the absence of Hg (II), to maintain its own expression at certain level. | ||
| − | [[Image:MerR structure.jpg]] | + | [[Image:MerR-structure.jpg]] |
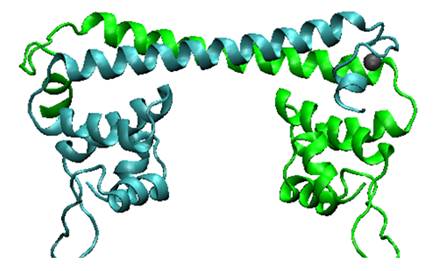
'''Fig.1. MerR dimer structure.''' We can see from its structure, upper part is the metal binding domain. When two monomers come together in the presence of mercury, to form a dimer, mercury ions will be captured inside. | '''Fig.1. MerR dimer structure.''' We can see from its structure, upper part is the metal binding domain. When two monomers come together in the presence of mercury, to form a dimer, mercury ions will be captured inside. | ||
Revision as of 17:53, 26 October 2010
RBS (B0034) + MerR (mercury-responsive transcription factor)
This part was designed as a translational unit for MerR expression.
The mercury resistance operon, mer, can enable bacteria to avoid and remove toxic metal Hg under the regulation of the MerR family transcriptional factor MerR. MerR acts as an activator of mer genes in response to the presence of Hg (II), while it will be reversed into a weak repressor in the absence of Hg (II), to maintain its own expression at certain level.
Fig.1. MerR dimer structure. We can see from its structure, upper part is the metal binding domain. When two monomers come together in the presence of mercury, to form a dimer, mercury ions will be captured inside.
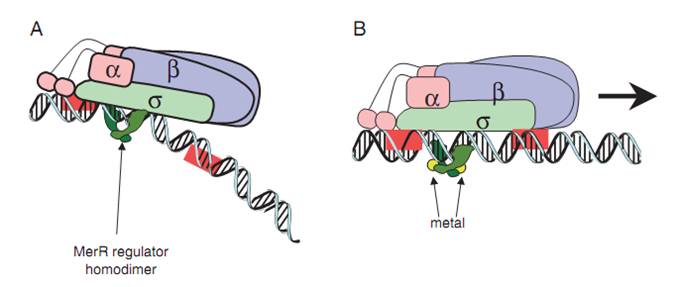
The MerR protein, in the form of homodimer, binds to the mer operon between the RNA polymerase binding sites of the promoter region. The attachment is independent on the presence of Hg(II). When the apo-MerR dimer bind to the dyad symmetrical operator DNA between the -35 and – 10 elements of mercury inducible promoter, PmerT, which has a unusually long spacer of 19 bp for MerR to bind on, the binding of RNA polymerase is inhibited. When Hg(II) is available in the environment, the ion binds to merR between the two subunits. The Hg-bound MerR can result in an a structural distortion of PmerT, allowing the RNA polymerase contacts to be made, leading to the expression of down-stream genes(Fig.2, Fig.3).
Fig.2. A generalized mechanism of MerR family regulator transcriptional activation. A: The dimeric MerR regulator binds to the operator region of the promoter and recruits RNA polymerase, forming a ternary complex. Transcription is slightly repressed because the apo-MerR regulator dimer has bent the promoter DNA such that RNA polymerase does not contact it properly. B: Upon binding the cognate metal ions (shown as cyan circles) the metallated MerR homodimer causes a realignment of the promoter such that RNA polymerase contacts the -35 and -10 sequences leading to open complex formation and transcription. Modified from Brown et al.
Fig.3. The model for the interaction of MerR and Hg(II) and its role in controlling PmerT transcription. Adapted from Jon L. Hobman, John Wilkie & Nigel L. Brown,2005. A: RNA polymerase (RNAP) transcribes merR from PmerR. MerR binds to the mer promoter/operator region (merOP) as a homodimer, recruits RNA polymerase, and represses transcription of merTPAD from PmerT. B: Hg(II) enters the bacterial cell by diffusion through the outer membrane, cytoplasm and inner membrane, and binds to three cysteine residues in the apo-MerR homodimer. The Hg-bound MerR homodimer causes an underwinding of merOP DNA, allowing RNAP to proceed with transcription of the resistance genes.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]