Difference between revisions of "Part:BBa K404201"
VolkerMorath (Talk | contribs) |
VolkerMorath (Talk | contribs) (→The Z34C motif (Z34C)) |
||
| Line 126: | Line 126: | ||
====The Z34C motif (Z34C)==== | ====The Z34C motif (Z34C)==== | ||
| − | + | [[Image:Freiburg10_Structures of Z34C Z38 and the Z-Domain.png|thumb|right|500px|Structural representaion of the Z38 and the Z34C motif]] | |
| + | |||
| + | The idea of this targeting approach is to utilise a minimized fragment of the [http://www.ncbi.nlm.nih.gov/protein/153106 Staphylococcal Protein A] that was first described in ''Staphylococcus aureus''. This gram-positive bacteria has evolved the 508 amino acid long protein A that has a high affinity for the Fc-domain of antibodies to protect itself from the immune systeme. Binding to the constant region of the antibodies is accomplished by the [http://www.ncbi.nlm.nih.gov/protein/1Q2NA Z-Domain] of Protein A that is 58-59 amino acids long, has alone a high affinity (Kd= 14,9 nM) for the antibodies and a three-helix bundle structure. In [http://www.ncbi.nlm.nih.gov/pubmed/8650153 [Braisted & Wells; 1996] ]the authors reduced the secundary structure to an two-helix bundle. This size reduction has lead to an drastic reduction of the affinity for IgG (>10^5 fold) which could be recovered by 13 amino acid exchanges resulting in a 38 amino acid long peptide with an satisfying affinity for IgG (Kd = 185 nM) termed [http://www.ncbi.nlm.nih.gov/protein/1ZDAA Z38]. This binding domain was subsequently improoved in [http://www.ncbi.nlm.nih.gov/pubmed/9294166 [Starovasnik et al.; 1997] ] by the insertion of a disulfide bridge connecting the ends of the helices leading to the binding domain [http://www.ncbi.nlm.nih.gov/protein/1ZDCA Z34C] which shows an increased affinity for IgG (Kd = 20 nM).<br> | ||
| + | This engineered antibody binding domain of 34 amino acids was then inserted into capsids of different viral vectors amongst others also the AAV. In [[Media:Freiburg10_Adeno-associated virus capsids displaying immunoglobulin-binding domains permit antibody-mediated vector retargeting to specific cell surface receptors.pdf| [Ried et al.; 2002] ]] the [http://www.ncbi.nlm.nih.gov/protein/1ZDCA Z34C] domain was inserted at position 587 into the capsid of the AAV resulting in viral vector that can be targeted to different target cells without genetic engineering. This targeting approach was then improved in [http://www.ncbi.nlm.nih.gov/pubmed/15922956 [Gigout et al.; 2005]] by the creation of mosaic vectors that contain only ~25% of recombinant VP-Proteins what resulted in 4 to 5 orders of magnitude more infectiosity compared to all-mutant viruses. | ||
| + | |||
<ins>References:</ins> | <ins>References:</ins> | ||
| − | + | *[http://www.ncbi.nlm.nih.gov/pubmed/8650153 [Braisted & Wells; 1996]] | |
| + | *[http://www.ncbi.nlm.nih.gov/pubmed/9294166 [Starovasnik et al.; 1997]] | ||
| + | *[http://www.ncbi.nlm.nih.gov/pubmed/11932421 [Ried et al.; 2002]] | ||
| + | *[http://www.ncbi.nlm.nih.gov/pubmed/15922956 [Gigout et al.; 2005]] | ||
====Risk assessment and terms of use==== | ====Risk assessment and terms of use==== | ||
Revision as of 22:45, 1 September 2010
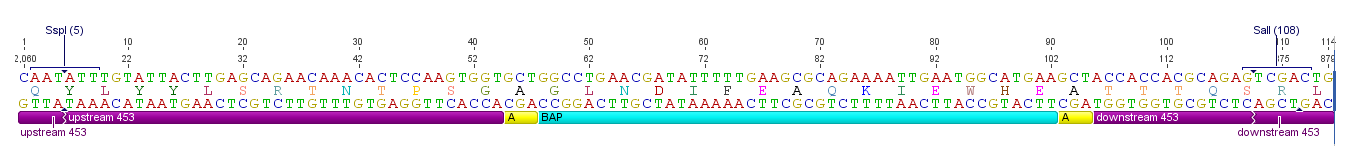
ViralBrick-453-BAP
| ViralBrick-453-BAP | |
|---|---|
| BioBrick Nr. | BBa_K404201 |
| RFC standard | RFC 25RFC 10 |
| Requirement | backbone without SspI, SalI, BamHI and PvuII |
| Source | synthetic |
| Submitted by | [http://2010.igem.org/Team:Freiburg_Bioware FreiGEM 2010] |
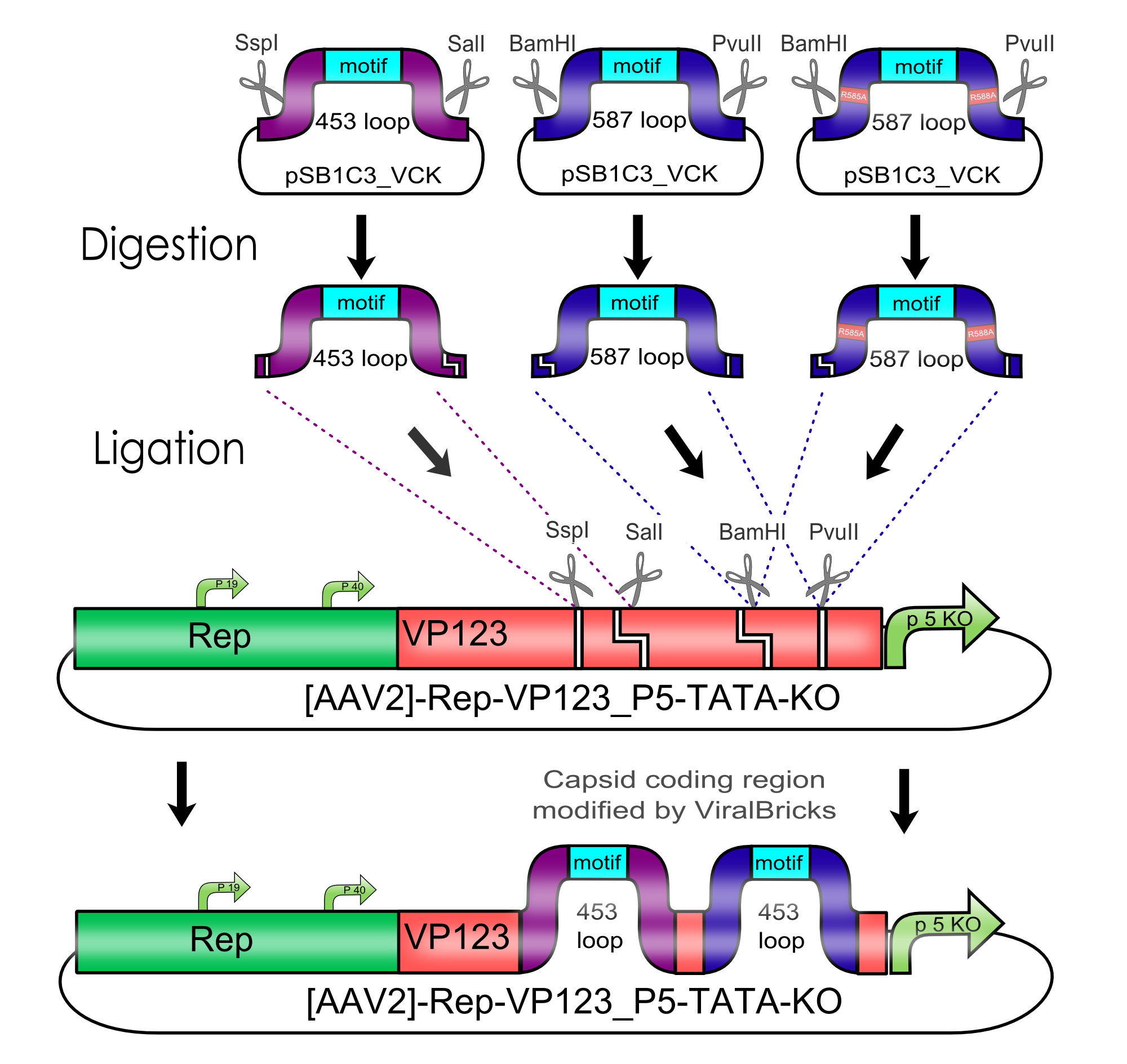
All capsid coding parts (e.g. (AAV2)-RepVP123) of the Virus Construction Kit designed by the iGEM Team Freiburg contain single cutting restriction sites on the side of the sequences coding for the two major surface exposed loops.
Using these restriction sites it is possible to insert functional motifs into the coding sequence for the viral capsid.
This BioBrick contains one of these functional motifs fanked by bilateral linkers and the viral sequences that code for the loop.
This category of BioBricks was termed ViralBrick to clarify that cloning does not function with the usual iGEM RFC restriction sites but with the mentioned ViralBrick restriction sites.
The four restriction sites for the ViralBrick insertion were designed in a way that the amino acid sequence of the viral capsid could be absolutely conserved. This was advisable because even slight changes in the viral capsid lead to drastically changed interactions with cellular surface receptors. For this reason we decided to insert these restriction sites in stead of using BioBrick assembly that would define at least two amino acids in the viral loop.
Modification of the Viral Capsid of the AAV2 using Viral Bricks
For therapeutical applications in human gene transfer the broad tropism for heparan sulfate proteoglycan (HSPG) has to be knocked-out and a novel tropism has to be inserted.
This retargeting can be realized either by insertion of functional motifs into the two major surface exposed loops or by fusion of these motifs to the N-terminus of the viral coat proteins.
The graphic on the right shows parts of the three-dimensional structure of a viral coat protein. The parts of the loop regions that are coded in the ViralBricks are shown in purple for the 453 loop and in blue for the 587 loop.
Cloning of Viral Bricks into capsid coding parts
In order to make loop insertions more convenient the following restriction sites were inserted into all capsid coding parts and already existing restriction sites were removed from the constructs.
The choice of these restriction sites was reasoned by enzyme performance, buffer compabilities and the number of existing restriction sites that had to be removed at other positions.
All restriction endonucleases were purchased from [http://www.neb-online.de NEB].
The 453 insertion site
References:
The 587 insertion site
References:
The Biotinylation Activating Peptide (BAP)
References:
The Histidine affinity Tag (His-tag)
References:
The RGD motif (BAP)
References:
The Z34C motif (Z34C)
The idea of this targeting approach is to utilise a minimized fragment of the [http://www.ncbi.nlm.nih.gov/protein/153106 Staphylococcal Protein A] that was first described in Staphylococcus aureus. This gram-positive bacteria has evolved the 508 amino acid long protein A that has a high affinity for the Fc-domain of antibodies to protect itself from the immune systeme. Binding to the constant region of the antibodies is accomplished by the [http://www.ncbi.nlm.nih.gov/protein/1Q2NA Z-Domain] of Protein A that is 58-59 amino acids long, has alone a high affinity (Kd= 14,9 nM) for the antibodies and a three-helix bundle structure. In [http://www.ncbi.nlm.nih.gov/pubmed/8650153 [Braisted & Wells; 1996] ]the authors reduced the secundary structure to an two-helix bundle. This size reduction has lead to an drastic reduction of the affinity for IgG (>10^5 fold) which could be recovered by 13 amino acid exchanges resulting in a 38 amino acid long peptide with an satisfying affinity for IgG (Kd = 185 nM) termed [http://www.ncbi.nlm.nih.gov/protein/1ZDAA Z38]. This binding domain was subsequently improoved in [http://www.ncbi.nlm.nih.gov/pubmed/9294166 [Starovasnik et al.; 1997] ] by the insertion of a disulfide bridge connecting the ends of the helices leading to the binding domain [http://www.ncbi.nlm.nih.gov/protein/1ZDCA Z34C] which shows an increased affinity for IgG (Kd = 20 nM).
This engineered antibody binding domain of 34 amino acids was then inserted into capsids of different viral vectors amongst others also the AAV. In [Ried et al.; 2002] the [http://www.ncbi.nlm.nih.gov/protein/1ZDCA Z34C] domain was inserted at position 587 into the capsid of the AAV resulting in viral vector that can be targeted to different target cells without genetic engineering. This targeting approach was then improved in [http://www.ncbi.nlm.nih.gov/pubmed/15922956 [Gigout et al.; 2005]] by the creation of mosaic vectors that contain only ~25% of recombinant VP-Proteins what resulted in 4 to 5 orders of magnitude more infectiosity compared to all-mutant viruses.
References:
- [http://www.ncbi.nlm.nih.gov/pubmed/8650153 [Braisted & Wells; 1996]]
- [http://www.ncbi.nlm.nih.gov/pubmed/9294166 [Starovasnik et al.; 1997]]
- [http://www.ncbi.nlm.nih.gov/pubmed/11932421 [Ried et al.; 2002]]
- [http://www.ncbi.nlm.nih.gov/pubmed/15922956 [Gigout et al.; 2005]]
Risk assessment and terms of use
Viral Systems based on the Adeno-associated Virus of the serotype 2 are to handle under Biosafety level 1 according to the german [http://bundesrecht.juris.de/gentg/index.html Act on Genetic Engineering] and the specification of the ZKBS on working with the AAV. This general classification is restricted to experiments that use the ITRs (Inverted Terminal Repeats) of the AAV2 and do not contain hazzardous gene sequences on the vector plasmid.
Please consider special provisions of law in your contry before using parts that contribute the production of genetically modified viral vectors.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]