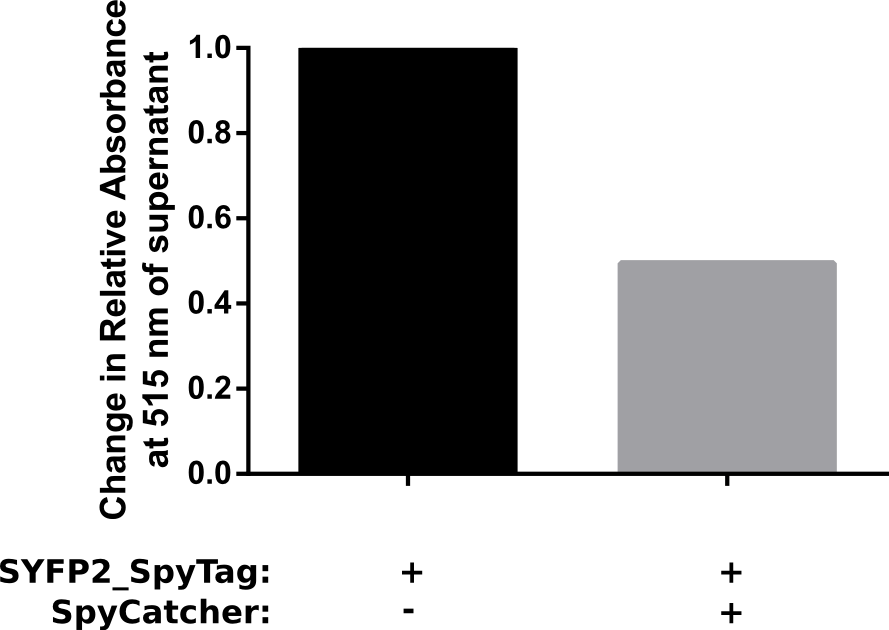Difference between revisions of "Part:BBa K1159200"
(→Contribution of iGEM Team LCG-china 2024) |
|||
| Line 114: | Line 114: | ||
</html> | </html> | ||
We have developed a new platform for cellulose modification using this system. This platform serves as a foundational framework, allowing for functionalization of cellulose while decoupling protein functionality from its compatibility with cellulose. This innovation enables future applications in cellulose modification without concerns about binding capabilities. | We have developed a new platform for cellulose modification using this system. This platform serves as a foundational framework, allowing for functionalization of cellulose while decoupling protein functionality from its compatibility with cellulose. This innovation enables future applications in cellulose modification without concerns about binding capabilities. | ||
| + | |||
| + | === Reference === | ||
| + | [1] Khatri, V., Jafari, M., Gaudreault, R., et al., 2023. Bionanocomposites with enhanced physical properties from curli amyloid assemblies and cellulose nanofibrils. Biomacromolecules, 24(11), pp.5290-5302. doi:10.1021/acs.biomac.3c00786.<br> | ||
| + | [2] Hatlem, D., Trunk, T., Linke, D. & Leo, J.C., 2019. Catching a SPY: using the SpyCatcher-SpyTag and related systems for labeling and localizing bacterial proteins. International Journal of Molecular Sciences, 20(9), p.2129. Published on 30 April 2019. doi:10.3390/ijms20092129.<br> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 11:23, 1 October 2024
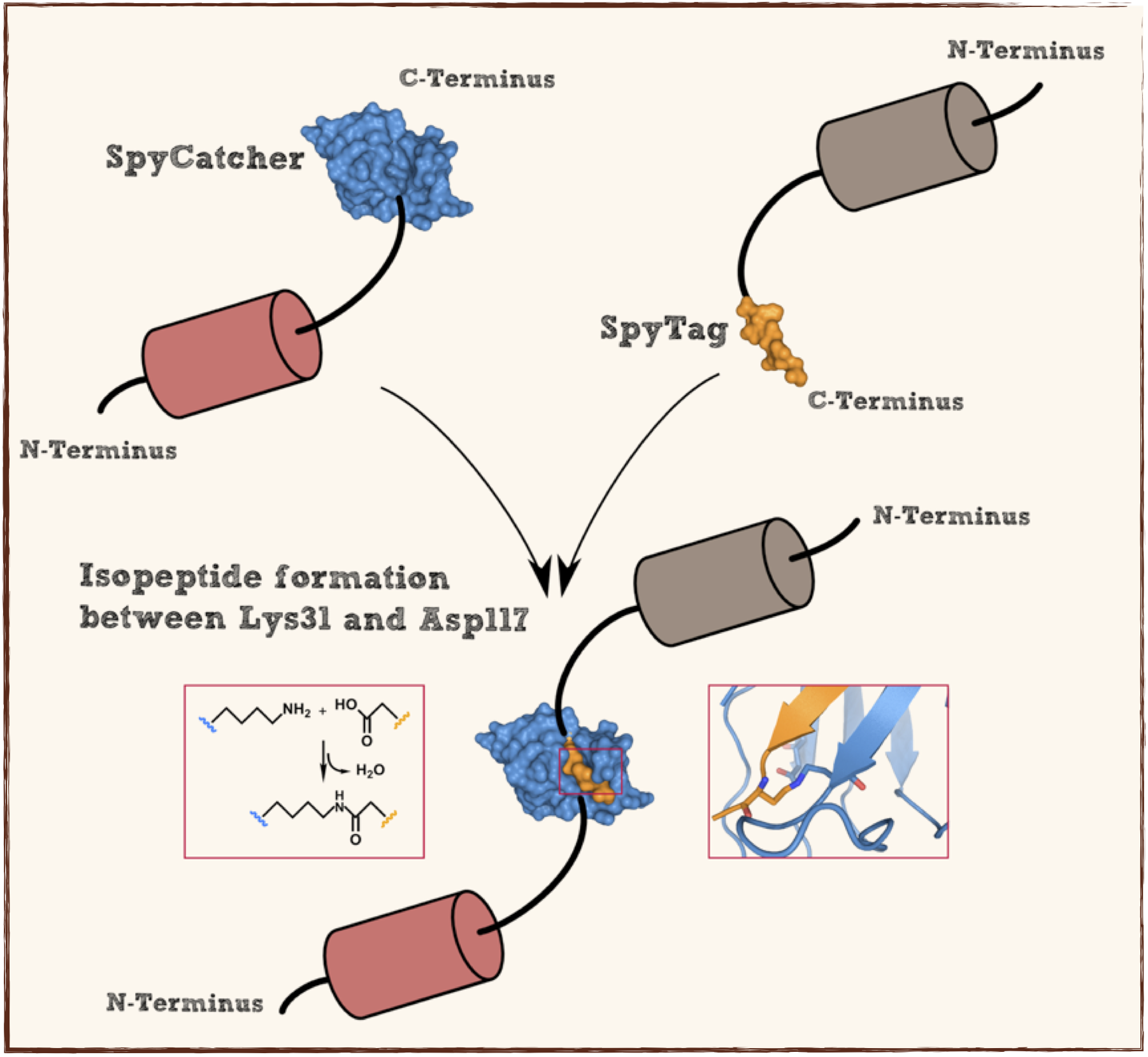
Splitted and engineered N-terminal FbaB for isopeptide bound formation (SpyCatcher) in RFC[25]
| Improved Part | |
|---|---|
| BioBrick Nr. | BBa_K2273015 |
| Submitted by | [http://2017.igem.org/Team:TU_Dresden TU Dresden] |
This part codes for a protein that recognizes and forms a covalent isopeptide bound to a oligopeptide. That oligopeptide can be found under BBa_K1159201. In contrast to the original SpyCatcher this part only contains the essential part for forming isopeptide bounds and does not contain a N-terminal His-tag and TEV cleavage site. Furthermore two amino acids were exchanged to improve the function, Ile34 was replaced by Glu34 which enhances the reaction rate, also the exposed hydrophobic redidue Met69 was replaced by Tyr69. This part is flanked by RFC[25] pre- and suffix for further protein fusions.
Experimental Evidence
Proper funtion of the construct was verified in a pulldown assay using 10 μM of SpyCatcher with N-terminal HisTag which was incubated with 5 μM of C-terminal spytagged SYFP2 for 1 h in PBS. Afterwards HisTag beads were added to bind the fusion product, resulting in a decrease of spytagged SYFP2 in the supernatant. The results of this assay are shown in Figure 2. The results show that after one hour at room temeperature, a two fold change in relative absorbance was observed. This verifies that the construct is working as intended.
Furthermore the two interaction partners were mixed, incubated and analyzed via SDS Page (reducing conditions) and coomassie staining.
Improvment
Inspired by the self-catalysis, Peking iGEM 2016 fused three SpyTag spaced by (VPGVG)4 with 6xHistag in N-terminal and other functional protein modules in C-terminal. The network forming capacity is not affected. Moreover, E.coli has successfully secreted our fused protein when a few of specific signal peptides are added.
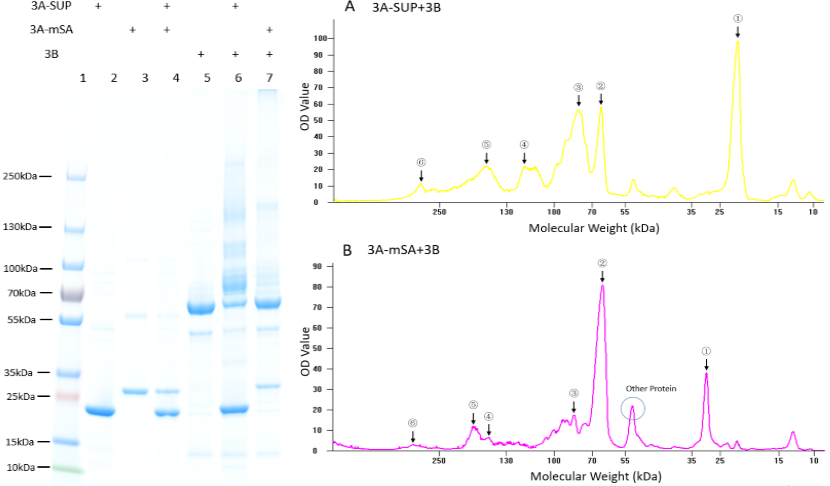
Fig. 1. Exploration of the polymerization ability of the 3A-SUP/3A-mSA with 3B. Left: the Coomassie Blue staining gel of basic experiment, which illustrates the basic cross-linking ability of 3A-SUP/3A-mSA and 3B. Lane 1, Thermo® Protein Marker; lane 2, 3A-SUP; lane 3, 3A-mSA; lane 4, 3A-SUP+3A-mSA; lane5, 3B; lane 6, 3A-SUP+3B; lane 7, 3A-mSA+3B. (Molecular Weight: 3A-SUP, 21.4kDa; 3A-mSA, 25.4kDa; 3B, 55.4(62) kDa. Right: A, the OD value of the lane 5 of oligomers produced by the mix of 3A-SUP and 3B. Peaks illustrate the monomers and the possible products, ①, 3A-SUP (21.4kDa); ②, 3B (62kDa); ③, 1x 3A-SUP+1x 3B (83.4kDa); ④, 3x 3A-SUP+1x 3B (126.2kDa); ⑤, 2x 3A-SUP+2x 3B (166.8kDa); ⑥, 4x 3A-SUP+4x 3B (333.6kDa). B, the OD value of the lane 6 of oligomers produced by the mix of 3A-mSA and 3B. Peaks illustrate the monomers and the possible products, ①, 3A-mSA (25.4kDa); ②, 3B (62kDa); ③, 1x 3A-mSA+1x 3B (87.4kDa); ④, 1x 3A-mSA+2x 3B (149.4kDa); ⑤, 2x 3A-mSA+2x 3B (174.4kDa); ⑥, 3x 3A-mSA+4x 3B (324.2kDa). (The software lane 1D was used to draw the graph.) (“3A-SUP” stands for “Triple SpyTag-SUP”, “3A-mSA” for “Triple SpyTag-mSA”, and “3B” is the abbreviation of “Triple SpyCatcher”)
We found that some new bands appeared above the band of 3B when it was mixed with monomers containing A, which demonstrated that our idea of forming functional hydrogel was executable. The products were mainly oligomers (Fig. 2 A-F, Table 1), for it is easy to form loops, which hindered the linkage between different monomers at such low concentration. Interestingly, with the restriction that A is equal to B in number, and the content of 3B was constant initially, the crosslinking ability at low concentration of these monomers were different with each other by comparing the surplus content of 3B. What’s more, it was surprising that the position of 3B band (~62kDa) was not accord with its theoretical weight (55.4kDa). Further experiments will be done to understand the differences.
If you want to learn more about Peking’s polymer network and the role of SUP in this network, please click here https://parts.igem.org/Part:BBa_K1989000", https://parts.igem.org/Part:BBa_K1989001" or https://parts.igem.org/Part:BBa_K1989002".
Reference
http://www.ncbi.nlm.nih.gov/pubmed/22366317 Zakeri et al., 2012 Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A. 20;109(12)
Characterization by iGEM Kyoto 2019
We measured the time development of SpyCatcher-SpyTag bond formation. Here, we show the time development of bond formation between SpyCatcher and "SpyTag inserted TmEncapsulin". SpyTag inserted TmEncapsulin(BBa_K3185000) is one of our basic protein-coding part. SpyTag inserted TmEncapsulin forms capsule, Virus-like particle, spontaneously as 60mer[1]. In this part, SpyTag is exposed to the surface of the protein capsule. So we can display SpyCatcher-fused protein on the capsule.[2]
In the experiment, an equal amount of SpyCatcher protein (SpyC) and SpyTag inserted TmEncapsulin (SpyTmEnc) were mixed and incubated at room temperature. At different time points, 10 min, 30 min, 60 min, 180 min, 360 min, 1200 min, the reaction was stopped by adding 2x SDS sample buffer. Mixed samples were assessed with SDS-PAGE. The intensities of the conjugated bands were quantified.
As shown in Fig. 1, conjugated bands become evident gradually (labeled with arrow). Signals were quantified and summarized in Fig. 2. The reaction looks saturated after 360 minutes, even though substrates still remain a lot. This might be explained by water evaporation while incubation. Otherwise, it is possible that a bound protein prevents another protein from binding to near sites on a capsule cage. If it is the case, it might limit the number of binding proteins on a capsule.

Reference
1 Putri, R.M., Allende-Ballestero, C., Luque, D., Klem, R., Rousou, K.A., Liu, A., Traulsen, C.H.H., Rurup, W.F., Koay, M.S.T., Castón, J.R., et al. (2017).
Structural Characterization of Native and Modified Encapsulins as Nanoplatforms for in Vitro Catalysis and Cellular Uptake.
ACS Nano 11, 12796–12804.
2 Bae, Y., Kim, G.J., Kim, H., Park, S.G., Jung, H.S., and Kang, S. (2018).
Engineering Tunable Dual Functional Protein Cage Nanoparticles Using Bacterial Superglue.
Biomacromolecules 19, 2896–2904.
Characterization by iGEM Team Keystone 2023
We incorporate the SpyCatcher-SpyTag covalent bonding system in our surface display system for displaying hpCA on the cell surface of Synechocystis PCC680. This system is essential for our project since it maximizes the binding of the carbonic anhydrase to substrate while ensuring its stability.
SpyCatcher-SpyTag is a versatile system which is known, among other applications, for the formation of multi-protein complexes. Here, we present a system in which SpyTag is fused to the native surface-exposed proteins Slp (BBa_K4863002) and PilA1 (BBa_K4863001) on Synechocystis PCC680 for surface display, and a purified hpCA-SpyCatcher (BBa_K4863005) complex produced by E. Coli BL21 (DE3) is mixed with the engineered Synechocystis to bind to SpyTag displayed on the cell surface. Functional surface display of hpCA is successfully achieved this way.
To test for the functionality of this surface display system, we mix a purified sfGFP-SpyCatcher complex with Synechocystis PCC 6803 expressing the protein complexes SpyTag-Slp and PilA1-SpyTag. If SpyTag is successfully displayed, the sfGFP-SpyCatcher complex will fuse with the surface displayed SpyTag via covalent bonding and the organism will obtain green florescence due to sfGFP. Florescence intensity of the supernatant of SpyTag-Slp expressing Synechocystis has significant difference compared to the WildType control and florescence intensity of the supernatant of PilA1-SpyTag expressing Synechocystis show some difference, demonstrating that some sfGFP have binded to the cell surface via bonding between SpyCatcher and SpyTag.

Fig.1: Florescence intensity of supernatant (absorbance wavelength: 488mm; excitation wavelength: 512mm). 1A: Florescence of Control (no bacteria added), WildType, and PilA1-SpyTag. 1B: Florescence of Control, WildType, and SpyTag-Slp.
For further verification, the cells were observed under a 40X florescent microscope. Clear green florescence signals show on the cell surface of <ir>Synechocystis</it> expressing SpyTag-Slp and few green florescence signals show on the cell surface of Synechocystis expressing PilA1-SpyTag, verifying successful display of SpyTag on the cell surface and successful fusion of the protein complex to the displayed SpyTag.

Therefore, we demonstrate successful surface display of SpyTag through fusion to both Slp and PilA1 and show that the SpyTag-SpyCatcher system can be applied for the surface display of a large protein on the cell surface of Synechocystis.
Contribution of iGEM Team LCG-china 2024
We have selected the Part BBa_K1159200 to complement existing data by adding new test results. Using the Co-Immunoprecipitation (Co-IP) method, we successfully verified the strong binding between SpyCatcher and SpyTag by demonstrating the formation of a complex, which confirms their interaction under physiological conditions. This approach provides additional binding data for the SpyTag system and also validates the feasibility of this system from an immunological perspective.




Reference
[1] Khatri, V., Jafari, M., Gaudreault, R., et al., 2023. Bionanocomposites with enhanced physical properties from curli amyloid assemblies and cellulose nanofibrils. Biomacromolecules, 24(11), pp.5290-5302. doi:10.1021/acs.biomac.3c00786.
[2] Hatlem, D., Trunk, T., Linke, D. & Leo, J.C., 2019. Catching a SPY: using the SpyCatcher-SpyTag and related systems for labeling and localizing bacterial proteins. International Journal of Molecular Sciences, 20(9), p.2129. Published on 30 April 2019. doi:10.3390/ijms20092129.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCGTTGATACC ... GCTCATATTACCGGT ORF from nucleotide position -8 to 345 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
Alignments (obtained from PredictProtein.org)
| ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
Subcellular Localization (reliability in brackets)
| Gene Ontology (reliability in brackets)
| |||||||||||||||||||||||||||||||||||||||||||||
Predicted features:
| ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
This part has been improved:BBa_K2273015