Difference between revisions of "Part:BBa K3113007"
SarahiGEMMUC (Talk | contribs) |
|||
| Line 13: | Line 13: | ||
The members of the L7Ae/15.5kD protein family that characteristically recognize K-turn motifs found in both archaeal and eukaryotic RNAs. In Archaea, the L7Ae protein uniquely binds the K-turn motif found in box C/D.<ref>Gagnon, Keith T et al. “Signature amino acids enable the archaeal L7Ae box C/D RNP core protein to recognize and bind the K-turn RNA motif.” RNA (New York, N.Y.) vol. 16,1 (2010): 79-90. doi:10.1261/rna.1692310</ref> | The members of the L7Ae/15.5kD protein family that characteristically recognize K-turn motifs found in both archaeal and eukaryotic RNAs. In Archaea, the L7Ae protein uniquely binds the K-turn motif found in box C/D.<ref>Gagnon, Keith T et al. “Signature amino acids enable the archaeal L7Ae box C/D RNP core protein to recognize and bind the K-turn RNA motif.” RNA (New York, N.Y.) vol. 16,1 (2010): 79-90. doi:10.1261/rna.1692310</ref> | ||
The K-turn is a ubiquitous structural motif in RNA forming a very tight kink in the axis of helical RNA that plays an important role in many aspects of RNA function. L7Ae is a member of a superfamily of proteins that bind K-turns in RNA, stabilizing the tightly kinked conformation. They are extremely widespread and are important in the assembly of RNA–protein complexes central to translation, splicing and site-specific RNA modification. <ref>Lilley, D. M. J. (2019). "The L7Ae proteins mediate a widespread and highly functional protein–RNA interaction." The Biochemist 41(2): 40-44.</ref> | The K-turn is a ubiquitous structural motif in RNA forming a very tight kink in the axis of helical RNA that plays an important role in many aspects of RNA function. L7Ae is a member of a superfamily of proteins that bind K-turns in RNA, stabilizing the tightly kinked conformation. They are extremely widespread and are important in the assembly of RNA–protein complexes central to translation, splicing and site-specific RNA modification. <ref>Lilley, D. M. J. (2019). "The L7Ae proteins mediate a widespread and highly functional protein–RNA interaction." The Biochemist 41(2): 40-44.</ref> | ||
| + | ==Literature Characterization by AFCM-Egypt== | ||
| + | NORD27 is a class of c/d box, and the study used a soluble nuclear fraction where fibrillarin is not detected and a portion of SNORD27 is present. | ||
| + | <html><div align="center"style="border:solid #17252A; width:50%;float:center;"><img style=" max-width:850px; | ||
| + | width:75%; | ||
| + | height:auto; | ||
| + | position: relative; | ||
| + | top: 50%; | ||
| + | left: 35%; | ||
| + | transform: translate( -50%); | ||
| + | padding-bottom:25px; | ||
| + | padding-top:25px; | ||
| + | "src="https://static.igem.wiki/teams/4586/wiki/literature-characterisation-parts/cd-box.png"> | ||
| + | <p class=MsoNormal align=center style='text-align:left;border:none;width:98% ;justify-content:center;'><span | ||
| + | lang=EN style='font-size:11.0pt;line-height:115%'>((A) The diagram represents HeLa cell fractionation under the original circumstances. (b) It is Western blot analysis, as 5% of the total volume of each fraction prepared was analyzed by Western blot-specific antibodies used to assess The components of spliceosomes (PRP8 and PRPF39), splicing regulators (CBP80/NCBP1 and hnRNPG), and constitutive SNORD binding proteins (NHP2L1/15.5K, NOP56, NOP58, and fibrillarin) (c) The arrow indicates SNORD27, and boxes refer to hosting exons. RPA was used to assess the distribution of SNORD27 in cellular fractions. The RPA probe covered the specified SNORD27 sequence as well as 5 nucleotides from the next intron. (d) Nuclear supernatant (C) was separated on a 10-45% glycerol gradient, which was subdivided into 20 fractions, and then RPA with an SNORD27 antisense probe was used to analyze and isolate RNA from 50% of each fraction. (E) Western blotting used antibodies recognizing markers for pre-mRNA processing (CBP80, hnRNPG, and SRSF6) to assess individual glycerol gradient fractions. (F) RPA determined The distribution of U1, U2, and U4 snRNAs in the fractions obtained by HeLa cell fractionation (A) (G)SNORD27's association with naturally occurring SNORD-binding proteins NHP2L1, NOP58, and NOP56 that were flag-tagged as well as endogenous fibrillarin were immunoprecipitated and purified together. Then, RT-PCR was used to find SNORD27. GFP-transfected cells were employed as a control. | ||
| + | </span></p></div></html> | ||
Latest revision as of 19:54, 11 October 2023
C/D-Box
C/D box is a stemloop structure shown to associate with four evolutionary conserved and essential proteins including the archaeal homolog (L7Ae) of Snu13.
Usage
The informational readout of ALiVE is RNA. To load a specific RNA into the vesicles we fused it to RNA-motifs which are bound specifically by RNA binding proteins. Two combinations of binding protein and motif were tested. One was the archaeal RNA binding protein L7Ae which binds to the K-turns in the box C/D snoRNP.
Biology
The members of the L7Ae/15.5kD protein family that characteristically recognize K-turn motifs found in both archaeal and eukaryotic RNAs. In Archaea, the L7Ae protein uniquely binds the K-turn motif found in box C/D.[1] The K-turn is a ubiquitous structural motif in RNA forming a very tight kink in the axis of helical RNA that plays an important role in many aspects of RNA function. L7Ae is a member of a superfamily of proteins that bind K-turns in RNA, stabilizing the tightly kinked conformation. They are extremely widespread and are important in the assembly of RNA–protein complexes central to translation, splicing and site-specific RNA modification. [2]
Literature Characterization by AFCM-Egypt
NORD27 is a class of c/d box, and the study used a soluble nuclear fraction where fibrillarin is not detected and a portion of SNORD27 is present.

((A) The diagram represents HeLa cell fractionation under the original circumstances. (b) It is Western blot analysis, as 5% of the total volume of each fraction prepared was analyzed by Western blot-specific antibodies used to assess The components of spliceosomes (PRP8 and PRPF39), splicing regulators (CBP80/NCBP1 and hnRNPG), and constitutive SNORD binding proteins (NHP2L1/15.5K, NOP56, NOP58, and fibrillarin) (c) The arrow indicates SNORD27, and boxes refer to hosting exons. RPA was used to assess the distribution of SNORD27 in cellular fractions. The RPA probe covered the specified SNORD27 sequence as well as 5 nucleotides from the next intron. (d) Nuclear supernatant (C) was separated on a 10-45% glycerol gradient, which was subdivided into 20 fractions, and then RPA with an SNORD27 antisense probe was used to analyze and isolate RNA from 50% of each fraction. (E) Western blotting used antibodies recognizing markers for pre-mRNA processing (CBP80, hnRNPG, and SRSF6) to assess individual glycerol gradient fractions. (F) RPA determined The distribution of U1, U2, and U4 snRNAs in the fractions obtained by HeLa cell fractionation (A) (G)SNORD27's association with naturally occurring SNORD-binding proteins NHP2L1, NOP58, and NOP56 that were flag-tagged as well as endogenous fibrillarin were immunoprecipitated and purified together. Then, RT-PCR was used to find SNORD27. GFP-transfected cells were employed as a control.
Characterization
qPCR
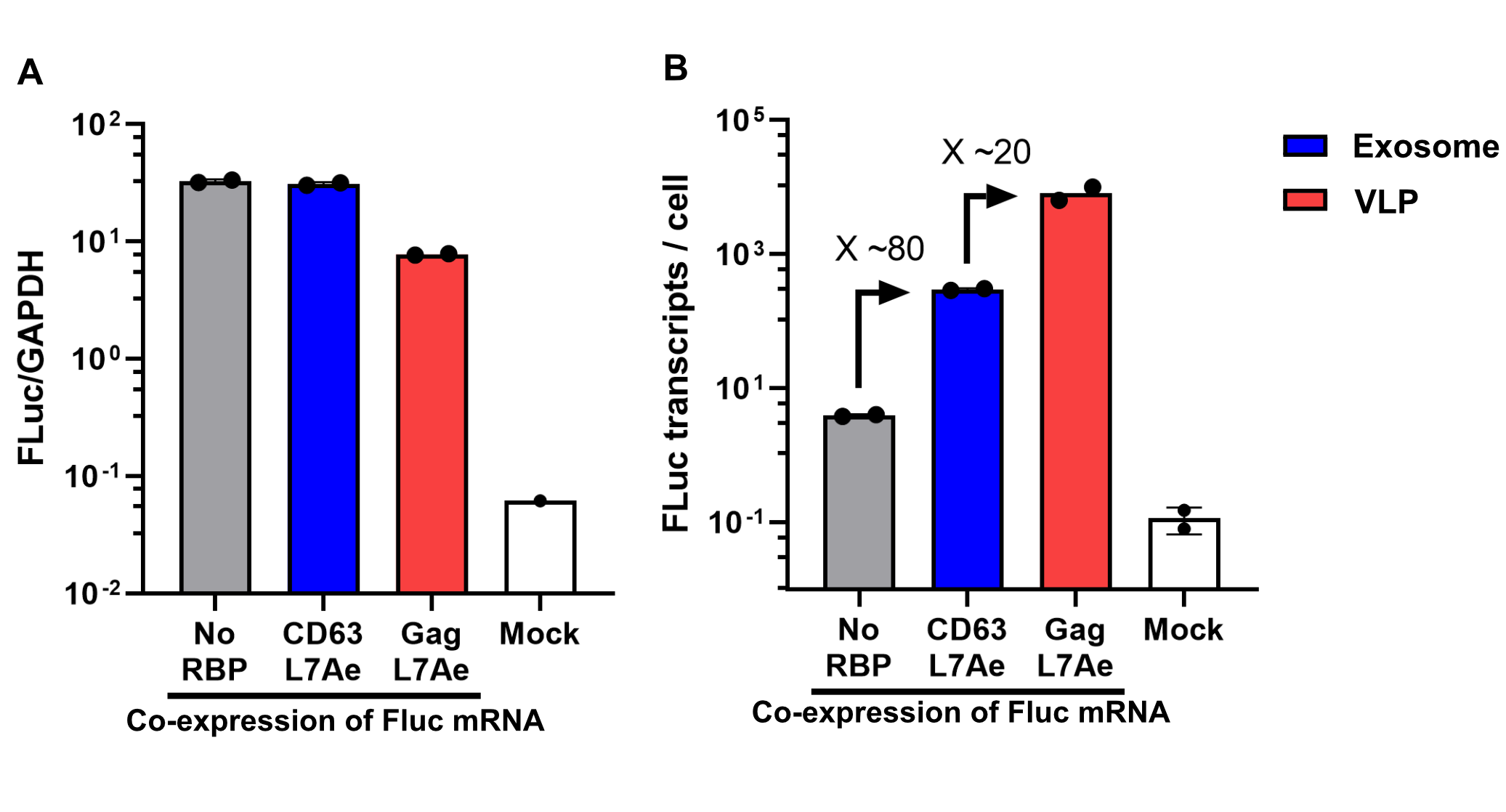
Using a coiled-coil system allowed us to successfully load the two different RNA binding proteins (RBPs) L7Ae or MCP into exosomes to export FLuc mRNA from HEK293T cells. We used the published parallel heterodimer pair P9SN:P10SN (Ljubetič et al. 2017), where P9SN was fused to the C-terminus of the exosome marker protein CD63 and P10SN N-terminal to the RNA binding proteins L7Ae or MCP. The export efficiencies of FLuc mRNA cargo measured by qPCR proved that coiled-coil mediated targeting of the RBPs L7Ae and MCP into exosomes worked. Exosomes formed from CD63-P9SN and loaded with P10SN-L7Ae or -MCP could export about 800 and 400 transcripts per cell, respectively. This also shows that the two RBPs have different export capabilities, with L7Ae exporting twice as many transcripts per cell compared to MCP.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
- ↑ Gagnon, Keith T et al. “Signature amino acids enable the archaeal L7Ae box C/D RNP core protein to recognize and bind the K-turn RNA motif.” RNA (New York, N.Y.) vol. 16,1 (2010): 79-90. doi:10.1261/rna.1692310
- ↑ Lilley, D. M. J. (2019). "The L7Ae proteins mediate a widespread and highly functional protein–RNA interaction." The Biochemist 41(2): 40-44.
