Difference between revisions of "Part:BBa K176015"
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K176015 short</partinfo> | <partinfo>BBa_K176015 short</partinfo> | ||
| Line 11: | Line 10: | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K176015 SequenceAndFeatures</partinfo> | <partinfo>BBa_K176015 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | =='''MEASUREMENT'''== | ||
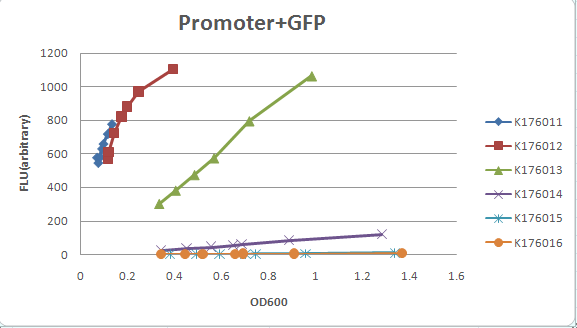
| + | We measured five '''constitutive promoter family member''' ( J23100, J23101, J23103, J23107, J23109, J23115 )strength by ligated each with a | ||
| + | GFP gene ( I13504) , and then moniter the OD600 and GFP expression. | ||
| + | K176011: J23100 + GFP ( I13504 ) | ||
| + | K176012: J23101 + GFP ( I13504) | ||
| + | K176013: K176009( J23107) + GFP ( I13504) | ||
| + | K176014: K176008( J23115) + GFP ( I13504) | ||
| + | K176015: J23109 + GFP ( I13504) | ||
| + | K176016: J23103 + GFP ( I13504) | ||
| + | ===data=== | ||
| + | J23109 + GFP | ||
| + | {| {{table}} | ||
| + | {|class="sortable" style="border:3px solid black; width: 70%" align="center" | ||
| + | | align="center" style="background:#f0f0f0;"|'''COLONY NUMBER''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''T(min)''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''OD600''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''FLU(arbitrary)''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''ST.FLU(ustc_st.1)''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''FLU/OD600(arbitrary)''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''ST.FLU/OD600(ustc_st.1)''' | ||
| + | |- | ||
| + | | Ⅰ||0||0.383||0.355666667||1.421699911||0.928633594||3.712010211 | ||
| + | |- | ||
| + | | Ⅰ||30||0.495||0.509333333||2.035948888||1.028956229||4.113028057 | ||
| + | |- | ||
| + | | Ⅰ||60||0.592||0.614||2.454331055||1.037162162||4.145829485 | ||
| + | |- | ||
| + | | Ⅰ||90||0.703||0.786||3.141863533||1.118065434||4.469222664 | ||
| + | |- | ||
| + | | Ⅰ||120||0.746||0.989||3.953311748||1.325737265||5.299345507 | ||
| + | |- | ||
| + | | Ⅰ||150||0.958||1.309333333||5.233774367||1.366736256||5.463230028 | ||
| + | |- | ||
| + | | Ⅰ||180||1.338||2.187||8.742055402||1.634529148||6.533673694 | ||
| + | |- | ||
| + | | Ⅱ||0||0.294||0.229333333||0.916709971||0.780045351||3.118061124 | ||
| + | |- | ||
| + | | Ⅱ||30||0.399||0.344333333||1.376397383||0.86299081||3.449617502 | ||
| + | |- | ||
| + | | Ⅱ||60||0.491||0.427666667||1.709504204||0.871011541||3.481678623 | ||
| + | |- | ||
| + | | Ⅱ||90||0.581||0.572||2.286445217||0.984509466||3.93536182 | ||
| + | |- | ||
| + | | Ⅱ||120||0.625||0.729||2.914018467||1.1664||4.662429548 | ||
| + | |- | ||
| + | | Ⅱ||150||0.835||0.964333333||3.854712129||1.15489022||4.616421711 | ||
| + | |- | ||
| + | | Ⅱ||180||1.213||1.821666667||7.2817151||1.501786205||6.003062737 | ||
| + | |} | ||
| + | {{USTC//promoters}} | ||
Revision as of 07:13, 20 October 2009
pCon 0.04(J23109)->RBS+GFP+T
pCon 0.04(J23109)+I13504
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 705
MEASUREMENT
We measured five constitutive promoter family member ( J23100, J23101, J23103, J23107, J23109, J23115 )strength by ligated each with a GFP gene ( I13504) , and then moniter the OD600 and GFP expression. K176011: J23100 + GFP ( I13504 ) K176012: J23101 + GFP ( I13504) K176013: K176009( J23107) + GFP ( I13504) K176014: K176008( J23115) + GFP ( I13504) K176015: J23109 + GFP ( I13504) K176016: J23103 + GFP ( I13504)
data
J23109 + GFP
| COLONY NUMBER | T(min) | OD600 | FLU(arbitrary) | ST.FLU(ustc_st.1) | FLU/OD600(arbitrary) | ST.FLU/OD600(ustc_st.1) |
| Ⅰ | 0 | 0.383 | 0.355666667 | 1.421699911 | 0.928633594 | 3.712010211 |
| Ⅰ | 30 | 0.495 | 0.509333333 | 2.035948888 | 1.028956229 | 4.113028057 |
| Ⅰ | 60 | 0.592 | 0.614 | 2.454331055 | 1.037162162 | 4.145829485 |
| Ⅰ | 90 | 0.703 | 0.786 | 3.141863533 | 1.118065434 | 4.469222664 |
| Ⅰ | 120 | 0.746 | 0.989 | 3.953311748 | 1.325737265 | 5.299345507 |
| Ⅰ | 150 | 0.958 | 1.309333333 | 5.233774367 | 1.366736256 | 5.463230028 |
| Ⅰ | 180 | 1.338 | 2.187 | 8.742055402 | 1.634529148 | 6.533673694 |
| Ⅱ | 0 | 0.294 | 0.229333333 | 0.916709971 | 0.780045351 | 3.118061124 |
| Ⅱ | 30 | 0.399 | 0.344333333 | 1.376397383 | 0.86299081 | 3.449617502 |
| Ⅱ | 60 | 0.491 | 0.427666667 | 1.709504204 | 0.871011541 | 3.481678623 |
| Ⅱ | 90 | 0.581 | 0.572 | 2.286445217 | 0.984509466 | 3.93536182 |
| Ⅱ | 120 | 0.625 | 0.729 | 2.914018467 | 1.1664 | 4.662429548 |
| Ⅱ | 150 | 0.835 | 0.964333333 | 3.854712129 | 1.15489022 | 4.616421711 |
| Ⅱ | 180 | 1.213 | 1.821666667 | 7.2817151 | 1.501786205 | 6.003062737 |
For the details of the definition of ST.FLU and the unit ustc_st1, please click here.
plot
protocol
1. Streak a plate of the strain which contain one of the parts listed in pSB1A3 .
2. Inoculate two 3ml cultures of supplemented M9 Medium and antibiotic( Ampicillin 0.1mg/ml) with single colony from the plate.
3. Cultures were grown in test tubes(BIO BASIC INC.12ml Polypropylene Round-bottom Culture Tubes With Graduations And Dual Cap Cat.No:TD444) for 16hrs at 37℃ with shaking at 200rpm.
4. Cultures were diluted 1:100 into 3ml fresh medium and grown for 3hrs.
5. Measure the fluorescence(SHIMDZU SPECTROFLUOROPHOTOMETER RF-5301PC, 250ul quartz cell path length 10mm,501 nm excitation,514 nm emission,1.5nm slit width) and absorbance (HITACHI UV-VIS spectrophotometer U-2810 ,200ul quartz cell,path length 10mm,600nm,1.5 nm slit width) every 30 minutes in the next 4hrs.