Difference between revisions of "Part:BBa K176014"
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K176014 short</partinfo> | <partinfo>BBa_K176014 short</partinfo> | ||
| Line 11: | Line 10: | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K176014 SequenceAndFeatures</partinfo> | <partinfo>BBa_K176014 SequenceAndFeatures</partinfo> | ||
| + | =='''MEASUREMENT'''== | ||
| + | We measured five '''constitutive promoter family member''' ( J23100, J23101, J23103, J23107, J23109, J23115, )strength by ligated each with a | ||
| + | GFP gene ( I13504) , and then moniter the OD600 and GFP expression. | ||
| + | K176011: J23100 + GFP ( I13504 ) | ||
| + | K176012: J23101 + GFP ( I13504) | ||
| + | K176013: K176009( J23107) + GFP ( I13504) | ||
| + | K176014: K176008( J23115) + GFP ( I13504) | ||
| + | K176015: J23109 + GFP ( I13504) | ||
| + | K176016: J23103 + GFP ( I13504) | ||
| + | ===data=== | ||
| + | K176008+GFP | ||
| + | {| {{table}} | ||
| + | {|class="sortable" style="border:3px solid black; width: 70%" align="center" | ||
| + | | align="center" style="background:#f0f0f0;"|'''COLONY NUMBER''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''T(min)''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''OD600''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''FLU(arbitrary)''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''ST.FLU(ustc_st.1)''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''FLU/OD600(arbitrary)''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''ST.FLU/OD600(ustc_st.1)''' | ||
| + | |- | ||
| + | | Ⅰ||0||0.344||6.204666667||24.80180144||18.03682171||72.09826 | ||
| + | |- | ||
| + | | Ⅰ||30||0.451||9.088666667||36.32996229||20.15225425||80.55424012 | ||
| + | |- | ||
| + | | Ⅰ||60||0.555||11.31433333||45.22657926||20.38618619||81.489332 | ||
| + | |- | ||
| + | | Ⅰ||90||0.65||13.382||53.49162569||20.58769231||82.29480876 | ||
| + | |- | ||
| + | | Ⅰ||120||0.685||15.55033333||62.15906517||22.70121655||90.74316083 | ||
| + | |- | ||
| + | | Ⅰ||150||0.888||20.59866667||82.33867637||23.1966967||92.72373465 | ||
| + | |- | ||
| + | | Ⅰ||180||1.281||29.948||119.7105968||23.37861046||93.45089523 | ||
| + | |- | ||
| + | | Ⅱ||0||0.358||6.817666667||27.25213521||19.04376164||76.12328272 | ||
| + | |- | ||
| + | | Ⅱ||30||0.456||9.824||39.26929688||21.54385965||86.11687912 | ||
| + | |- | ||
| + | | Ⅱ||60||0.55||11.94633333||47.75286139||21.72060606||86.82338434 | ||
| + | |- | ||
| + | | Ⅱ||90||0.647||15.013||60.01119239||23.20401855||92.75300215 | ||
| + | |- | ||
| + | | Ⅱ||120||0.679||16.711||66.79857697||24.61119293||98.37787477 | ||
| + | |- | ||
| + | | Ⅱ||150||0.866||22.18966667||88.69835179||25.62317167||102.423039 | ||
| + | |- | ||
| + | | Ⅱ||180||1.247||31.647||126.5019787||25.37850842||101.445051 | ||
| + | |} | ||
| + | {{USTC//promoters}} | ||
Revision as of 07:07, 20 October 2009
pCon 0.15(K176008)->RBS+GFP+T
pCon 0.15(K176008)+I13504
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 705
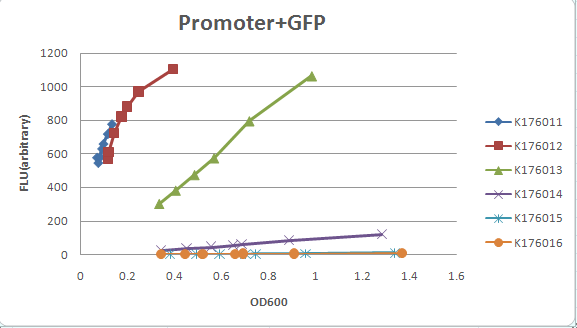
MEASUREMENT
We measured five constitutive promoter family member ( J23100, J23101, J23103, J23107, J23109, J23115, )strength by ligated each with a GFP gene ( I13504) , and then moniter the OD600 and GFP expression. K176011: J23100 + GFP ( I13504 ) K176012: J23101 + GFP ( I13504) K176013: K176009( J23107) + GFP ( I13504) K176014: K176008( J23115) + GFP ( I13504) K176015: J23109 + GFP ( I13504) K176016: J23103 + GFP ( I13504)
data
K176008+GFP
| COLONY NUMBER | T(min) | OD600 | FLU(arbitrary) | ST.FLU(ustc_st.1) | FLU/OD600(arbitrary) | ST.FLU/OD600(ustc_st.1) |
| Ⅰ | 0 | 0.344 | 6.204666667 | 24.80180144 | 18.03682171 | 72.09826 |
| Ⅰ | 30 | 0.451 | 9.088666667 | 36.32996229 | 20.15225425 | 80.55424012 |
| Ⅰ | 60 | 0.555 | 11.31433333 | 45.22657926 | 20.38618619 | 81.489332 |
| Ⅰ | 90 | 0.65 | 13.382 | 53.49162569 | 20.58769231 | 82.29480876 |
| Ⅰ | 120 | 0.685 | 15.55033333 | 62.15906517 | 22.70121655 | 90.74316083 |
| Ⅰ | 150 | 0.888 | 20.59866667 | 82.33867637 | 23.1966967 | 92.72373465 |
| Ⅰ | 180 | 1.281 | 29.948 | 119.7105968 | 23.37861046 | 93.45089523 |
| Ⅱ | 0 | 0.358 | 6.817666667 | 27.25213521 | 19.04376164 | 76.12328272 |
| Ⅱ | 30 | 0.456 | 9.824 | 39.26929688 | 21.54385965 | 86.11687912 |
| Ⅱ | 60 | 0.55 | 11.94633333 | 47.75286139 | 21.72060606 | 86.82338434 |
| Ⅱ | 90 | 0.647 | 15.013 | 60.01119239 | 23.20401855 | 92.75300215 |
| Ⅱ | 120 | 0.679 | 16.711 | 66.79857697 | 24.61119293 | 98.37787477 |
| Ⅱ | 150 | 0.866 | 22.18966667 | 88.69835179 | 25.62317167 | 102.423039 |
| Ⅱ | 180 | 1.247 | 31.647 | 126.5019787 | 25.37850842 | 101.445051 |
For the details of the definition of ST.FLU and the unit ustc_st1, please click here.
plot
protocol
1. Streak a plate of the strain which contain one of the parts listed in pSB1A3 .
2. Inoculate two 3ml cultures of supplemented M9 Medium and antibiotic( Ampicillin 0.1mg/ml) with single colony from the plate.
3. Cultures were grown in test tubes(BIO BASIC INC.12ml Polypropylene Round-bottom Culture Tubes With Graduations And Dual Cap Cat.No:TD444) for 16hrs at 37℃ with shaking at 200rpm.
4. Cultures were diluted 1:100 into 3ml fresh medium and grown for 3hrs.
5. Measure the fluorescence(SHIMDZU SPECTROFLUOROPHOTOMETER RF-5301PC, 250ul quartz cell path length 10mm,501 nm excitation,514 nm emission,1.5nm slit width) and absorbance (HITACHI UV-VIS spectrophotometer U-2810 ,200ul quartz cell,path length 10mm,600nm,1.5 nm slit width) every 30 minutes in the next 4hrs.