Difference between revisions of "Part:BBa K190015:Design"
m (→References) |
m |
||
| Line 12: | Line 12: | ||
[[Image:PBca1020-r0040.jpg|200px]] | [[Image:PBca1020-r0040.jpg|200px]] | ||
| + | |||
| + | ===Modelling=== | ||
| + | |||
| + | {{GraphHeader}} | ||
| + | <html> | ||
| + | <script type="text/javascript" src="/Team:Groningen/Modelling/Model.js?action=raw"></script> | ||
| + | <script type="text/javascript" src="/Team:Groningen/Modelling/Arsenic.js?action=raw"></script> | ||
| + | </html> | ||
| + | |||
| + | The three graphs below illustrate the promoter response after induction with arsenic (directly in the cell, with the equivalent of 1µM in the solution) with and without constitutive expression of ArsR (the first two graphs) and with slower production and degradation of ArsR (the two left graphs). Also, each graph has a line showing the formation of a product behind the ars promoter that does not degrade (and has production rate 1), subtracting the production that would have occurred without induction to show the effect of adding arsenic. Some conclusions: | ||
| + | |||
| + | * Constitutive expression of ArsR greatly reduces (and slows) the promoter response. | ||
| + | * On the other hand, if we divide the production and degradation rates of ArsR by ten the promoter response is ten times slower, producing ten times as much product. | ||
| + | * In the bottom-right graph the induction is done gradually (the amount of arsenic increases linearly during the first five minutes), showing the high-pass behaviour of the promoter and that this can negatively impact product formation. | ||
| + | |||
| + | <html> | ||
| + | <script type="text/javascript"> | ||
| + | addOnloadHook(computePromoterActivation); | ||
| + | |||
| + | function computePromoterActivation() { | ||
| + | // Set up constants | ||
| + | var maxt = 600; | ||
| + | var c = arsenicModelConstants(); | ||
| + | var cNP = {}, cS = {}, cG = {}; | ||
| + | c.v5 = 0; | ||
| + | c.k8 = 0; | ||
| + | c.pro = 0; | ||
| + | c.ars2T = 0; | ||
| + | for(var a in c) { | ||
| + | cNP[a] = c[a]; | ||
| + | cS[a] = c[a]; | ||
| + | cG[a] = c[a]; | ||
| + | } | ||
| + | |||
| + | var Vcell = 1 * 1e-15; // micrometer^3/cell -> liter/cell | ||
| + | var avogadro = 6.02214179e23; // 1/mol | ||
| + | c.pro = 2/(avogadro*Vcell); // 1/cell -> mol/L | ||
| + | cS.tauR *= 10; | ||
| + | cS.beta1 /= 10; | ||
| + | cS.beta3 /= 10; | ||
| + | cG.ars2T = 100*cG.ars1T; | ||
| + | |||
| + | // Initialize | ||
| + | var x0 = arsenicModelInitialization(c,0); | ||
| + | var xNP0 = arsenicModelInitialization(cNP,0); | ||
| + | var xS0 = arsenicModelInitialization(cS,0); | ||
| + | var x20 = arsenicModelInitialization(c,0); | ||
| + | var xG0 = arsenicModelInitialization(cG,0); | ||
| + | var AsT = 1e-6*c.Vs; | ||
| + | x0.AsinT = AsT/c.Vc; | ||
| + | xNP0.AsinT = AsT/c.Vc; | ||
| + | xS0.AsinT = AsT/c.Vc; | ||
| + | x20.AsinT = 0; | ||
| + | xG0.AsinT = AsT/c.Vc; | ||
| + | |||
| + | // Simulate | ||
| + | var x = simulate(x0,maxt,function(t,d){return arsenicModelGradient(c,d);}); | ||
| + | var xNP = simulate(xNP0,maxt,function(t,d){return arsenicModelGradient(cNP,d);}); | ||
| + | var xS = simulate(xS0,maxt*10,function(t,d){return arsenicModelGradient(cS,d);}); | ||
| + | var xG = simulate(xG0,maxt,function(t,d){return arsenicModelGradient(cG,d);}); | ||
| + | var x2 = simulate(x0,maxt,function(t,d){ | ||
| + | var Dx = arsenicModelGradient(c,d); | ||
| + | if (t<maxt/2) Dx.AsinT += (AsT/c.Vc)*2/maxt; | ||
| + | return Dx; | ||
| + | }); | ||
| + | |||
| + | // Output | ||
| + | function convertToSeries(c,x0,x) { | ||
| + | var bAsin, cAsin, ArsR, ars, arsP, arsE; | ||
| + | var arsInt = 0; | ||
| + | var series = [[],[]]; | ||
| + | var preTime = -x.time[x._arsF.length-1]/(60*20); | ||
| + | arsE = x0._arsF; | ||
| + | series[0].push({x:preTime,y:100*arsE}); | ||
| + | series[0].push({x:0,y:100*arsE}); | ||
| + | series[1].push({x:preTime,y:0}); | ||
| + | for(var i=0; i<x._arsF.length; i++) { | ||
| + | ars = x._arsF[i]; | ||
| + | if (i>0) arsInt += (x.time[i]-x.time[i-1])*(ars+arsP)/2; | ||
| + | series[0].push({x:x.time[i]/60,y:100*ars}); | ||
| + | series[1].push({x:x.time[i]/60,y:(arsInt-x.time[i]*arsE)}); | ||
| + | arsP = ars; | ||
| + | } | ||
| + | return series; | ||
| + | } | ||
| + | document.getElementById("promoterActivationData").data = { | ||
| + | ars:convertToSeries(c,x0,x), | ||
| + | arsNP:convertToSeries(cNP,xNP0,xNP), | ||
| + | arsS:convertToSeries(cS,xS0,xS), | ||
| + | arsG:convertToSeries(cG,xG0,xG), | ||
| + | ars2:convertToSeries(c,x20,x2)}; | ||
| + | var graphNodes = [document.getElementById("promoterActivationGraph"), | ||
| + | document.getElementById("promoterActivationGraphNP"), | ||
| + | document.getElementById("promoterActivationGraphS"), | ||
| + | document.getElementById("promoterActivationGraphG"), | ||
| + | document.getElementById("promoterActivationGraph2")]; | ||
| + | for(var i in graphNodes) if (graphNodes[i]) graphNodes[i].refresh(); | ||
| + | } | ||
| + | </script> | ||
| + | </html> | ||
| + | <span id="promoterActivationData"></span> | ||
| + | {| | ||
| + | |{{graph|Team:Groningen/Graphs/PromoterActivationNP|promoterActivitationGraphNP}} | ||
| + | |{{graph|Team:Groningen/Graphs/PromoterActivation|promoterActivitationGraph}} | ||
| + | |{{graph|Team:Groningen/Graphs/PromoterActivationG|promoterActivitationGraphG}} | ||
| + | |- | ||
| + | |{{graph|Team:Groningen/Graphs/PromoterActivationSlow|promoterActivitationGraphS}} | ||
| + | |{{graph|Team:Groningen/Graphs/PromoterActivation2|promoterActivitationGraph2}} | ||
| + | |} | ||
===Source=== | ===Source=== | ||
Revision as of 04:48, 20 October 2009
Arsenic Promoter (ArsR regulated)
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Design Notes
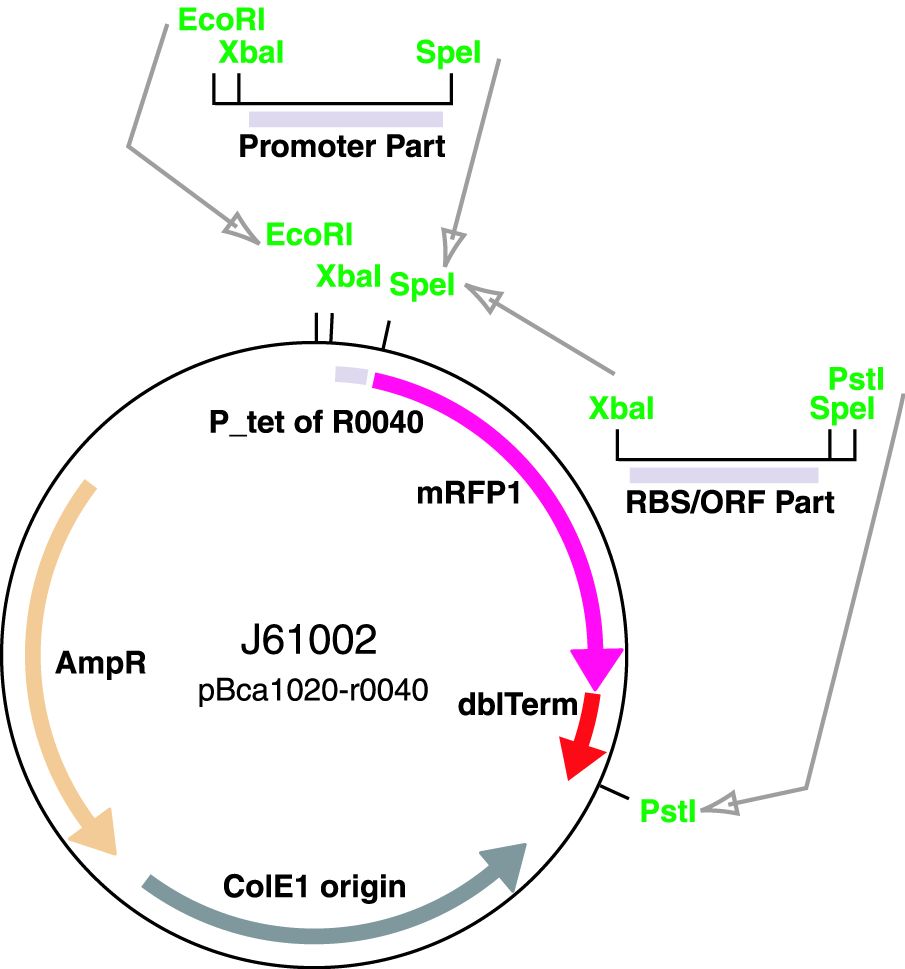
The ArsR regulated promoter has been cloned into the BBa_J61002-R0040 plasmid for construction of promoter basic parts and their derivatives.
Insertion of a promoter element between the XbaI and SpeI sites resulted in a RFP reporter while retaining the ability to do biobrick assembly.
Modelling
The three graphs below illustrate the promoter response after induction with arsenic (directly in the cell, with the equivalent of 1µM in the solution) with and without constitutive expression of ArsR (the first two graphs) and with slower production and degradation of ArsR (the two left graphs). Also, each graph has a line showing the formation of a product behind the ars promoter that does not degrade (and has production rate 1), subtracting the production that would have occurred without induction to show the effect of adding arsenic. Some conclusions:
- Constitutive expression of ArsR greatly reduces (and slows) the promoter response.
- On the other hand, if we divide the production and degradation rates of ArsR by ten the promoter response is ten times slower, producing ten times as much product.
- In the bottom-right graph the induction is done gradually (the amount of arsenic increases linearly during the first five minutes), showing the high-pass behaviour of the promoter and that this can negatively impact product formation.
Loading graph...
|
Loading graph...
|
Loading graph...
|
Loading graph...
|
Loading graph...
|
Source
E.coli TOP10
References
1) C. Xu, W. Shi, and B.P. Rosen (1996) The Chromosomal arsR Gene of Escherichia coli Encodes a trans-acting Metalloregulatory Protein, The Journal of Biological Chemistry, Vol. 271, No. 5, Issue of February 2, pp. 2427–2432.