Difference between revisions of "Part:BBa K4275042"
(→Characterisation of the Cellulosome Complex) |
|||
| Line 4: | Line 4: | ||
This composite part is designed for the expression of secondary scaffoldin of the cellulosome complex with the scaffoldin part modified to contain 3 type II cohesin domain and had its anchoring domain changed to an antigen domain. The protein is paired with strong promoter and strong terminator tested suitable for the expression of the coding sequence. The fusion protein is expressed and the antigen domain can interact with nanobody domain to use in cell surface display and the type II cohesin domain can fix the primary scaffoldin subunits to construct cellulosome complexes[1]. | This composite part is designed for the expression of secondary scaffoldin of the cellulosome complex with the scaffoldin part modified to contain 3 type II cohesin domain and had its anchoring domain changed to an antigen domain. The protein is paired with strong promoter and strong terminator tested suitable for the expression of the coding sequence. The fusion protein is expressed and the antigen domain can interact with nanobody domain to use in cell surface display and the type II cohesin domain can fix the primary scaffoldin subunits to construct cellulosome complexes[1]. | ||
| + | |||
==Usage and Biology== | ==Usage and Biology== | ||
| Line 16: | Line 17: | ||
<h3>Mini-scaffold construction of cellulosome</h3> | <h3>Mini-scaffold construction of cellulosome</h3> | ||
| − | We constructed E.coli expression vectors for the mini-scaffold protein subunits. The scaffoldin components of the wild-type cellulosome subunits are large protein scaffolds that would bring a massive protein burden to the bacterial host secreting them. We modified the coding sequences for the wild-type cellulosome protein scaffold, as shown in (Fig.1A and Fig.1B) | + | We constructed <i>E.coli</i> expression vectors for the mini-scaffold protein subunits. The scaffoldin components of the wild-type cellulosome subunits are large protein scaffolds that would bring a massive protein burden to the bacterial host secreting them. We modified the coding sequences for the wild-type cellulosome protein scaffold, as shown in (Fig.1A and Fig.1B) |
The mini-scaffolds were successfully expressed by our host, verified by the SDS-PAGE analysis shown in (Fig.1C and Fig.1D) | The mini-scaffolds were successfully expressed by our host, verified by the SDS-PAGE analysis shown in (Fig.1C and Fig.1D) | ||
| Line 22: | Line 23: | ||
[[Image:GreatBay SCIE--Part Fig4.png|thumbnail|850px|center|'''Figure 1:''' | [[Image:GreatBay SCIE--Part Fig4.png|thumbnail|850px|center|'''Figure 1:''' | ||
| − | Mini-scaffold expression in E. coli BL21. (A) Construction of primary scaffold CipA1B2C (i.e., 1 CBM3 and 2 type I cohesin) (B) Construction of anchorage scaffold OlpB-Ag3 (i.e., 3 type II cohesin). (C) SDS-page analysis for CipA1B2C. (D) SDS-page analysis for OlpB-Ag3.]] | + | Mini-scaffold expression in <i>E.coli</i> BL21. (A) Construction of primary scaffold CipA1B2C (i.e., 1 CBM3 and 2 type I cohesin) (B) Construction of anchorage scaffold OlpB-Ag3 (i.e., 3 type II cohesin). (C) SDS-page analysis for CipA1B2C. (D) SDS-page analysis for OlpB-Ag3.]] |
| Line 30: | Line 31: | ||
In order to verify the three levels of protein-protein interaction that assembles our cellulosome complex, Ag3-eforRED, DocI-eforRED, and DocII-eforRED vectors were constructed and cultured for IPTG-inducible expression (Fig. 2D). SDS-page analysis was performed with lysed cells and all three targeted proteins were identified in both whole cell and supernatant (Fig. 3A and 3B). | In order to verify the three levels of protein-protein interaction that assembles our cellulosome complex, Ag3-eforRED, DocI-eforRED, and DocII-eforRED vectors were constructed and cultured for IPTG-inducible expression (Fig. 2D). SDS-page analysis was performed with lysed cells and all three targeted proteins were identified in both whole cell and supernatant (Fig. 3A and 3B). | ||
| − | The nanobody-antigen interaction was verified by mixing intact E. coli cells displaying Neae-Nb3 with the supernatant of Ag3-eforRED (Fig. 2A). Red fluorescent characteristics were observed in the pellets after resuspending the centrifuged mixture, which is absent in the control group that only contains Neae-Nb3 (Fig. 3C). | + | The nanobody-antigen interaction was verified by mixing intact <i>E.coli</i> cells displaying Neae-Nb3 with the supernatant of Ag3-eforRED (Fig. 2A). Red fluorescent characteristics were observed in the pellets after resuspending the centrifuged mixture, which is absent in the control group that only contains Neae-Nb3 (Fig. 3C). |
| − | After that, the type II cohesin-dockerin interaction was tested using the mixture of Neae-Nb3, OlpB-Ag3, and the type II | + | After that, the type II cohesin-dockerin interaction was tested using the mixture of Neae-Nb3, OlpB-Ag3, and the type II dockerin fused with eforRED (Fig. 2B). A negative control lacking OlpB-Ag3 was set up for result comparison. Centrifugation was used to remove supernatant and the red fluorescence was only identified in pellets of the sample group, confirming the type II cohesin-dockerin interaction (Fig. 3D). |
Finally, the association between type I cohesin and type I dockerin was validated using the mixture of Neae-Nb3, OlpB-Ag3, CipA1B2C, and DocI-eforRED (Fig. 2C), red fluorescence was detected in the resuspended mixture while it was not observed in the control group lacking the primary scaffold CipA1B2C (Fig. 3E), verifying the type I cohesin-dockerin interaction. | Finally, the association between type I cohesin and type I dockerin was validated using the mixture of Neae-Nb3, OlpB-Ag3, CipA1B2C, and DocI-eforRED (Fig. 2C), red fluorescence was detected in the resuspended mixture while it was not observed in the control group lacking the primary scaffold CipA1B2C (Fig. 3E), verifying the type I cohesin-dockerin interaction. | ||
| Line 45: | Line 46: | ||
[[Image:GreatBay SCIE--Part Fig6.png|thumbnail|850px|center|'''Figure 3:''' | [[Image:GreatBay SCIE--Part Fig6.png|thumbnail|850px|center|'''Figure 3:''' | ||
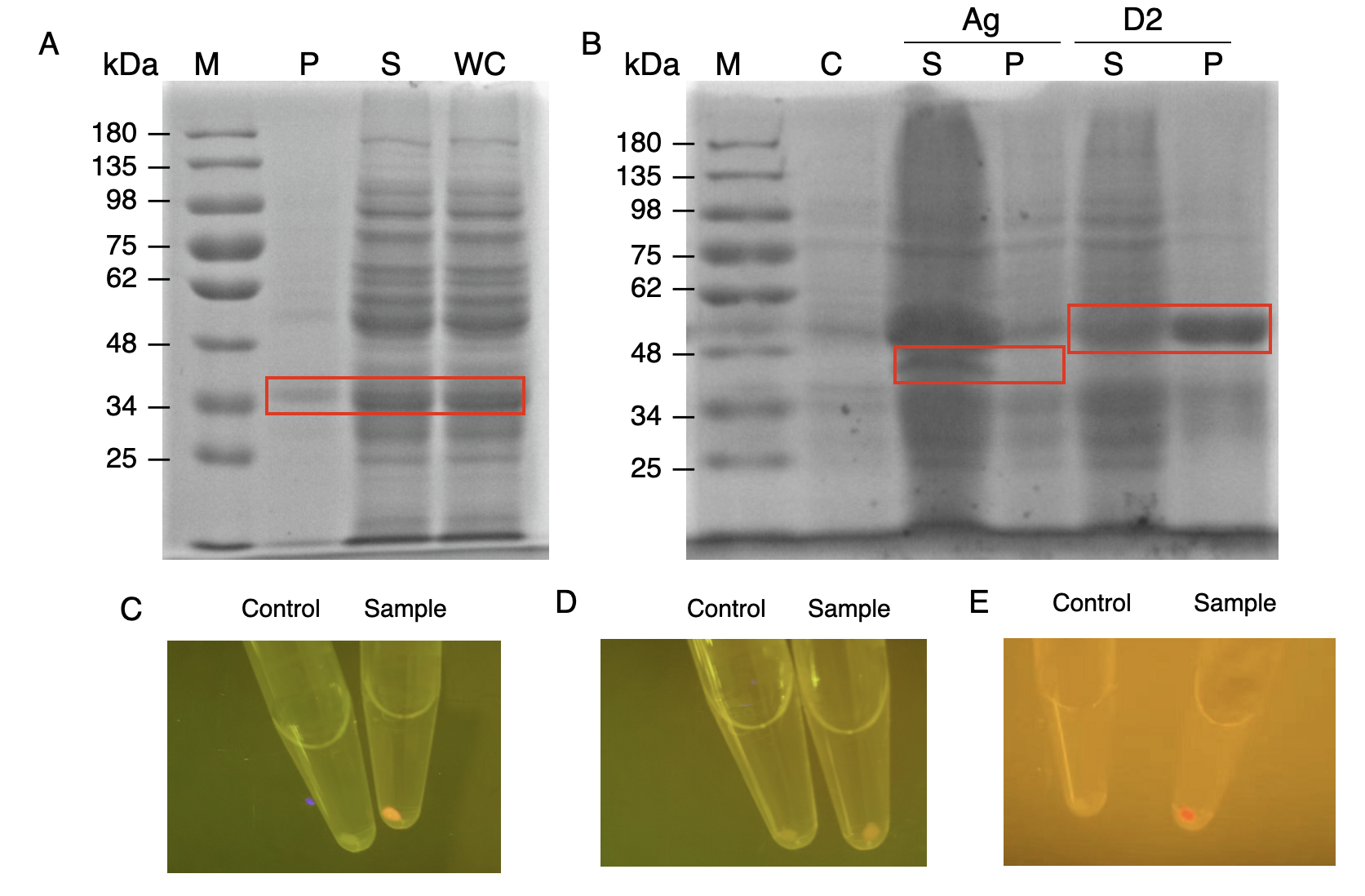
| − | Production and assay of the scaffold proteins (A) SDS-PAGE analysis for the presence of type I dockerin-eforRed (B) SDS-PAGE analysis of Ag3-eforRed and type II dockerin -eforRed containing type II dockerin fused with an eforRed domain (C) The fluorescence indication for antigen-nanobody interaction for E. | + | Production and assay of the scaffold proteins (A) SDS-PAGE analysis for the presence of type I dockerin-eforRed (B) SDS-PAGE analysis of Ag3-eforRed and type II dockerin -eforRed containing type II dockerin fused with an eforRed domain (C) The fluorescence indication for antigen-nanobody interaction for <i>E.coli</i> surface display with an Ag3 control group and a sample group (D) The fluorescence indication for type II cohesin-dockerin interactions with a type II dockerin control group and a sample group (E) The fluorescence indication for type I cohesin-dockerin interactions with a type I dockerin control group and a sample group.]] |
| Line 52: | Line 53: | ||
<h3>Cellulosome construction</h3> | <h3>Cellulosome construction</h3> | ||
| − | We assembled the cellulose-like complex on the surface of E.coli by adding primary scaffold proteins, cellulases and cellulase boosters onto E.coli expressing secondary scaffold proteins. The mixture was centrifuged and resuspended in tris-HCl. The mixture underwent centrifugation and resuspension using tris-HCl, and cellulose was added to the mixture. | + | We assembled the cellulose-like complex on the surface of <i>E.coli</i> by adding primary scaffold proteins, cellulases and cellulase boosters onto <i>E.coli</i> expressing secondary scaffold proteins. The mixture was centrifuged and resuspended in tris-HCl. The mixture underwent centrifugation and resuspension using tris-HCl, and cellulose was added to the mixture. |
After 24h, the mixture was filtered and tested for glucose by Benedict's test. From the result, we determined that the cellulosome-like complexes are able to degrade cellulose at a higher efficiency than cell-free cellulases mixture. | After 24h, the mixture was filtered and tested for glucose by Benedict's test. From the result, we determined that the cellulosome-like complexes are able to degrade cellulose at a higher efficiency than cell-free cellulases mixture. | ||
| Line 60: | Line 61: | ||
[[Image:GreatBay SCIE--Part Fig9.png|thumbnail|850px|center|'''Figure 4:''' | [[Image:GreatBay SCIE--Part Fig9.png|thumbnail|850px|center|'''Figure 4:''' | ||
The Benedict’s quantitative and qualitative tests for reducing sugar produced by the enzymatic or cellulosomal degradation of cellulose (A) Benedict’s qualitative test result for reducing sugar production through 24h of cellulose degradation by cellulosome, cellulosome without boosters, nanobody presenting cell+free cellulases+cellulase boosters, nanobody presenting cell+cellulases and nanobody presenting cell control from left to right (B) Benedict’s quantitative test for absorbance of the samples obtained from the Benedict’s qualitative test at 635 nm wavelength.]] | The Benedict’s quantitative and qualitative tests for reducing sugar produced by the enzymatic or cellulosomal degradation of cellulose (A) Benedict’s qualitative test result for reducing sugar production through 24h of cellulose degradation by cellulosome, cellulosome without boosters, nanobody presenting cell+free cellulases+cellulase boosters, nanobody presenting cell+cellulases and nanobody presenting cell control from left to right (B) Benedict’s quantitative test for absorbance of the samples obtained from the Benedict’s qualitative test at 635 nm wavelength.]] | ||
| + | |||
| + | |||
==Sequence and Features== | ==Sequence and Features== | ||
Latest revision as of 13:12, 13 October 2022
pT7-RBS-OlpB-Ag3-tT7
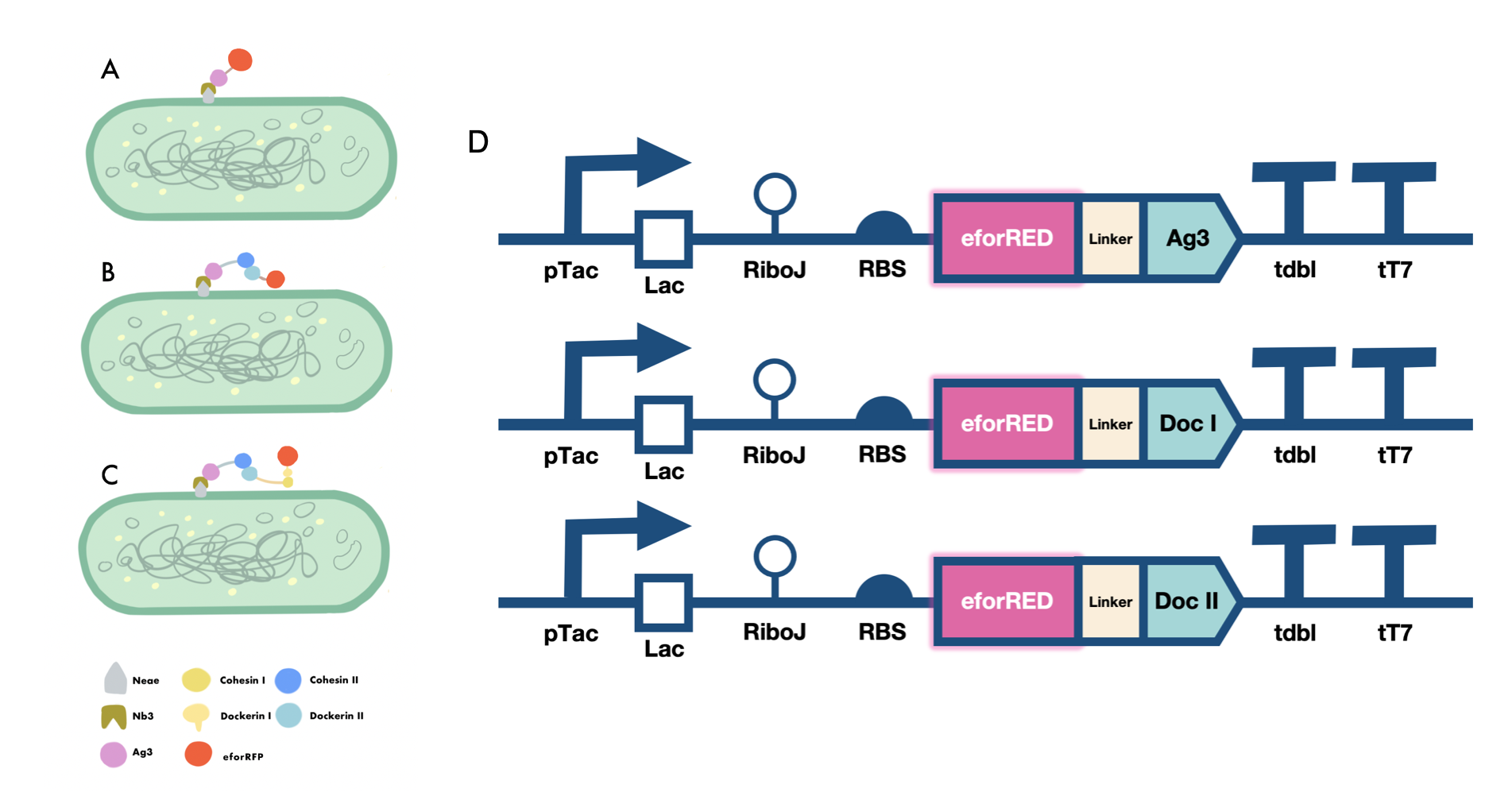
This composite part is designed for the expression of secondary scaffoldin of the cellulosome complex with the scaffoldin part modified to contain 3 type II cohesin domain and had its anchoring domain changed to an antigen domain. The protein is paired with strong promoter and strong terminator tested suitable for the expression of the coding sequence. The fusion protein is expressed and the antigen domain can interact with nanobody domain to use in cell surface display and the type II cohesin domain can fix the primary scaffoldin subunits to construct cellulosome complexes[1].
Usage and Biology
The protein can also be used in surface display by antigen-nanobody interactions because there is an antigen domain fused at C-terminus. The secondary scaffoldin subunit part contain multiple type II cohesin domain for the specific cohesin-dockerin interactions for fixing the primary scaffold components of the cellulosome complex for assembling of the cellulosome complex nanomachine.
Characterisation of the Cellulosome Complex
Mini-scaffold construction of cellulosome
We constructed E.coli expression vectors for the mini-scaffold protein subunits. The scaffoldin components of the wild-type cellulosome subunits are large protein scaffolds that would bring a massive protein burden to the bacterial host secreting them. We modified the coding sequences for the wild-type cellulosome protein scaffold, as shown in (Fig.1A and Fig.1B) The mini-scaffolds were successfully expressed by our host, verified by the SDS-PAGE analysis shown in (Fig.1C and Fig.1D)
Functionality testing of our mini-scaffold
In order to verify the three levels of protein-protein interaction that assembles our cellulosome complex, Ag3-eforRED, DocI-eforRED, and DocII-eforRED vectors were constructed and cultured for IPTG-inducible expression (Fig. 2D). SDS-page analysis was performed with lysed cells and all three targeted proteins were identified in both whole cell and supernatant (Fig. 3A and 3B).
The nanobody-antigen interaction was verified by mixing intact E.coli cells displaying Neae-Nb3 with the supernatant of Ag3-eforRED (Fig. 2A). Red fluorescent characteristics were observed in the pellets after resuspending the centrifuged mixture, which is absent in the control group that only contains Neae-Nb3 (Fig. 3C).
After that, the type II cohesin-dockerin interaction was tested using the mixture of Neae-Nb3, OlpB-Ag3, and the type II dockerin fused with eforRED (Fig. 2B). A negative control lacking OlpB-Ag3 was set up for result comparison. Centrifugation was used to remove supernatant and the red fluorescence was only identified in pellets of the sample group, confirming the type II cohesin-dockerin interaction (Fig. 3D).
Finally, the association between type I cohesin and type I dockerin was validated using the mixture of Neae-Nb3, OlpB-Ag3, CipA1B2C, and DocI-eforRED (Fig. 2C), red fluorescence was detected in the resuspended mixture while it was not observed in the control group lacking the primary scaffold CipA1B2C (Fig. 3E), verifying the type I cohesin-dockerin interaction.
Cellulosome construction
We assembled the cellulose-like complex on the surface of E.coli by adding primary scaffold proteins, cellulases and cellulase boosters onto E.coli expressing secondary scaffold proteins. The mixture was centrifuged and resuspended in tris-HCl. The mixture underwent centrifugation and resuspension using tris-HCl, and cellulose was added to the mixture.
After 24h, the mixture was filtered and tested for glucose by Benedict's test. From the result, we determined that the cellulosome-like complexes are able to degrade cellulose at a higher efficiency than cell-free cellulases mixture. The overall success in engineering our project was verified by the successful construction of cellulosome complex and degrading cellulose to reducing sugars.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 2058
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 746
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1292
Illegal NgoMIV site found at 1381
Illegal AgeI site found at 223
Illegal AgeI site found at 359
Illegal AgeI site found at 410
Illegal AgeI site found at 542
Illegal AgeI site found at 908
Illegal AgeI site found at 926
Illegal AgeI site found at 1352 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 1109
Illegal SapI.rc site found at 1121
Illegal SapI.rc site found at 1745
References
1. Anandharaj, Marimuthu et al. "Constructing A Yeast To Express The Largest Cellulosome Complex On The Cell Surface". Proceedings Of The National Academy Of Sciences, vol 117, no. 5, 2020, pp. 2385-2394. Proceedings Of The National Academy Of Sciences, https://doi.org/10.1073/pnas.1916529117.

