Difference between revisions of "Part:BBa K1529151"
Alexandra L (Talk | contribs) (→Biology) |
Alexandra L (Talk | contribs) (→Contribution From CAU_China 2022) |
||
| Line 17: | Line 17: | ||
'''Summary''': Added information about how this gene catalyze the formation of oxalic acid. | '''Summary''': Added information about how this gene catalyze the formation of oxalic acid. | ||
<br> | <br> | ||
| − | === | + | ===Biology=== |
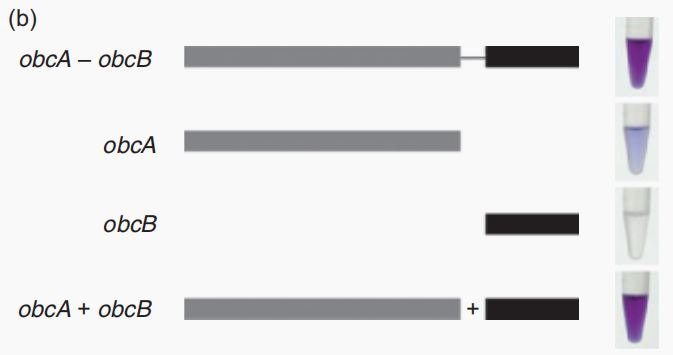
| − | <p>CAU_China 2022 | + | <p>CAU_China 2022 plan to use the CDS sequence of _obcB_ in this composite part and the expression product of _obcB_ is one of the main enzyme in in the oxalate secretion circuit<sup>[1]</sup>. It can act together with ObcA to catalyze oxaloacetic acid (OAA) and citric acid to form oxalic acid (OA). When there is only ObcB, it cannot form oxalic acid<sup>[2]</sup>.</p> |
| − | < | + | |
[[File:CAU-obcAB-pic.jpeg|600px|thumb|center|]] | [[File:CAU-obcAB-pic.jpeg|600px|thumb|center|]] | ||
<p style="text-align: center;"><b>Fig.1 Effects of <i>obcA</i> and <i>obcB</i>, the shade of purple reflects the concentration of oxalic acid<sup>[1]</sup>.</b></p > | <p style="text-align: center;"><b>Fig.1 Effects of <i>obcA</i> and <i>obcB</i>, the shade of purple reflects the concentration of oxalic acid<sup>[1]</sup>.</b></p > | ||
<br> | <br> | ||
| − | |||
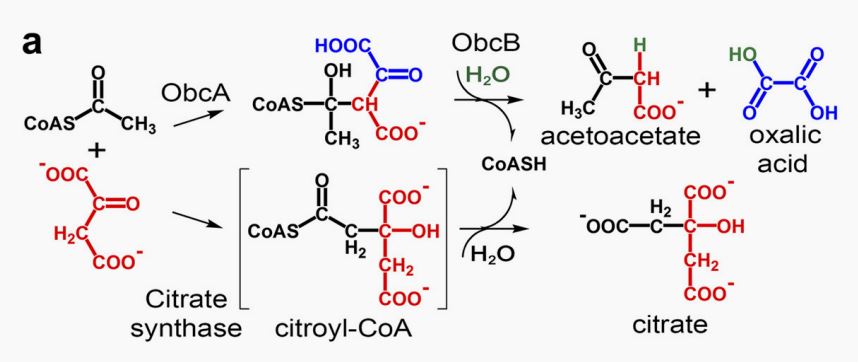
<p>When producing oxalic acid, <i>obcB</i> turns C6-CoA adduct into CoA, acetoacetate,and oxalic acid</p > | <p>When producing oxalic acid, <i>obcB</i> turns C6-CoA adduct into CoA, acetoacetate,and oxalic acid</p > | ||
<br> | <br> | ||
| Line 30: | Line 28: | ||
<p style="text-align: center;"><b>Fig.2 Mechanism of <i>obcA</i> and <i>obcB</i><sup>[2]</sup>.</b></p > | <p style="text-align: center;"><b>Fig.2 Mechanism of <i>obcA</i> and <i>obcB</i><sup>[2]</sup>.</b></p > | ||
| − | <p> | + | <p>Considering the importance of _obcA_ and _obcB_ occur togrther in the biosynthesis of oxalic acid, the CDS of them are in close proximity to each other, even encoded on a single polycistronic message. The implication of this for our project is that we can make these two genes share the same promoter<sup>[2]</sup>.</p> |
<br> | <br> | ||
[[File:CAU-obcAB-gene.jpeg|600px|thumb|center|]] | [[File:CAU-obcAB-gene.jpeg|600px|thumb|center|]] | ||
Revision as of 05:48, 7 October 2022
PT7_obcB
1
Contribution From CAU_China 2022
Group: CAU_China, 2022 https://2022.igem.org/Team:CAU_China
Author: Liu Muhua
Summary: Added information about how this gene catalyze the formation of oxalic acid.
Biology
CAU_China 2022 plan to use the CDS sequence of _obcB_ in this composite part and the expression product of _obcB_ is one of the main enzyme in in the oxalate secretion circuit[1]. It can act together with ObcA to catalyze oxaloacetic acid (OAA) and citric acid to form oxalic acid (OA). When there is only ObcB, it cannot form oxalic acid[2].
Fig.1 Effects of obcA and obcB, the shade of purple reflects the concentration of oxalic acid[1].
When producing oxalic acid, obcB turns C6-CoA adduct into CoA, acetoacetate,and oxalic acid
Fig.2 Mechanism of obcA and obcB[2].
Considering the importance of _obcA_ and _obcB_ occur togrther in the biosynthesis of oxalic acid, the CDS of them are in close proximity to each other, even encoded on a single polycistronic message. The implication of this for our project is that we can make these two genes share the same promoter[2].
Fig.3 Distribution of obcA and obcB[1].
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 669
- 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 669
Illegal NotI site found at 582
Illegal NotI site found at 687 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 650
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 669
- 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 669
Illegal NgoMIV site found at 460
Illegal NgoMIV site found at 501
Illegal NgoMIV site found at 625 - 1000COMPATIBLE WITH RFC[1000]
References
[1]Nakata, Paul A, and Cixin He. “Oxalic acid biosynthesis is encoded by an operon in Burkholderia glumae.” FEMS microbiology letters vol. 304,2 (2010): 177-82. doi:10.1111/j.1574-6968.2010.01895.x
[2]Oh, Juntaek et al. “Structural basis for bacterial quorum sensing-mediated oxalogenesis.” The Journal of biological chemistry vol. 289,16 (2014): 11465-11475. doi:10.1074/jbc.M113.543462