Difference between revisions of "Part:BBa K4304002"
| Line 11: | Line 11: | ||
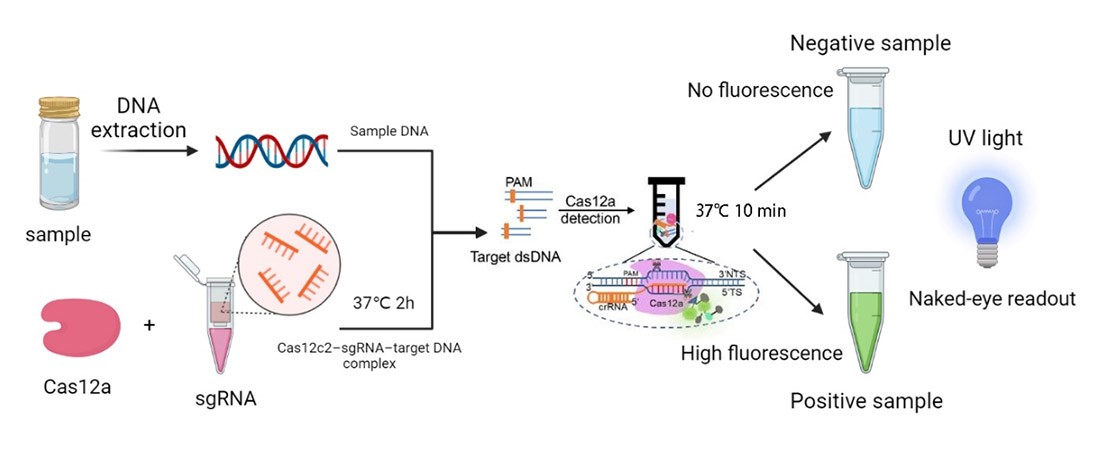
We synthesized four DNA fragments amplified from the pathogenic microorganisms we chose, and insert them in the pUC57 vector. Next, we purified the FnCas12a protein and synthesized probes with the 6-FAM flag. Then, we mixed those materials and the reaction buffer to develop the in vitro reaction system, and we also changed the concentration of the target genes. Finally, we detected the fluorescence of the reaction system and measured the activity of FnCas12a (Figure 1). | We synthesized four DNA fragments amplified from the pathogenic microorganisms we chose, and insert them in the pUC57 vector. Next, we purified the FnCas12a protein and synthesized probes with the 6-FAM flag. Then, we mixed those materials and the reaction buffer to develop the in vitro reaction system, and we also changed the concentration of the target genes. Finally, we detected the fluorescence of the reaction system and measured the activity of FnCas12a (Figure 1). | ||
[[File:T-- YkPaO--BBa K4304002-figure1.jpg|500px|thumb|center| Figure 1. The workflow of our project.]] | [[File:T-- YkPaO--BBa K4304002-figure1.jpg|500px|thumb|center| Figure 1. The workflow of our project.]] | ||
| + | |||
| + | ==Engineering Success == | ||
| + | |||
| + | FnCas12a, which is amplified from Francisella novicida, is a new class II family of CRISPR-Cas RNA-programmable endonucleases with unique features that make it a very attractive alternative or complement to Cas9 for genome engineering. | ||
| + | |||
| + | a) Construction of pathogenic micro-organisms expression plasmids | ||
| + | We designed 5 plasmids: the FnCas12 protein expression plasmid, 16S, cagA, ipaH, and invA expression plasmids. Among them, the DNA fragments 16S and cagA are amplified from the genome of Helicobacter Pylori, and the gene fragments ipaH and invA are amplified from salmonella and shigella genomic DNA respectively. | ||
| + | |||
| + | In order to construct our plasmids, we let the company synthesize the DNA fragments, FnCas12 was inserted into the pET28a vector, and the fragments 16S, cagA, ipaH, and invA were inserted into the pUC57 vector. The constructed plasmids were contained in E. coli strains, we streak inoculated them on LB solid medium plates containing corresponding antibiotics, and incubate them at 37℃ overnight. | ||
| + | |||
| + | |||
| + | b) Verification of the microorganisms expression plasmids | ||
| + | We used TAE agarose gel electrophoresis to testify the presence of oligo DNA in the plasmid by performing PCR and then doing gel electrophoresis of the amplicons (Figure 2). | ||
| + | [[File:T-- YkPaO--BBa K4304002-figure2.jpg|500px|thumb|center|Figure 2. Gel electrophoresis to verify the construction of oligo DNA containing plasmids.]] | ||
| + | |||
| + | Our results show that a band of 200bp to 400bp is present in cagA, 16S, invA, and ipaH, but not in negative control (NC) lanes. Because oligo DNA has a size between 200bp to 400bp, our result supports the fact that the plasmids contain desired oligo DNA. The four plasmid transformations were successful. | ||
| + | |||
| + | === Expression and purification of Cas12a protein=== | ||
| + | |||
| + | We transformed the pET28a-FnCas12a expression plasmid into E. coli BL21(DE3) competent cells, and cultured at 37℃ overnight (Figure 1A). we inoculated a single colony into LB (Kana+) culture medium, incubated overnight, and then transferred the cultured medium into 1L fresh LB (Kana+) culture medium. We induced the expression of FnCas12a with IPTG when the OD600 was around 0.6-1.0, and cultured at 16℃ for 12h. Subsequently, we used nickel affinity purification to purify the acquired Cas12a proteins from other proteins in E. coli (Figure 1B). | ||
| + | [[File:T--YkPaO--BBa K4304007-figure1.jpg|500px|thumb|center|Figure 3. Expression and purification of protein FnCas12a.]] | ||
| + | |||
| + | A. Incubate the plasmid pET28a-FnCas12a containing BL21(DE3). | ||
| + | B. SDS-PAGE electrophoresis gel of Cas12a protein compared to nonspecific protein impurities. | ||
| + | |||
| + | ===Bicinchoninic Acid Assay (BCA)=== | ||
| + | Cas12a protein has a size of 130kDa. The SDS-PAGE electrophoresis result indicates that the Cas12a protein is present in the solution we collected at 130kDa, and not present in the nonspecific protein impurities. Thus, Cas12a proteins were expressed and purified with high quality. | ||
| + | |||
| + | Then, we tested the concentration of Cas12a protein by Bicinchoninic Acid Assay (BCA), using SpectraMax i3x Multi-Mode Microplate Reader with the absorption peak at 562nm (Figure 2). | ||
| + | [[File:T--YkPaO--BBa K4304007-figure2.jpg|500px|thumb|center|Figure 4. BCA method standard linear regression line for calculation of protein concentration.]] | ||
| + | |||
Revision as of 08:43, 27 September 2022
cas12a-opt
Cas12a
Contribution
Based on CRISPR pathogenic microbial detection technology was developed in recent years, and was applied to a series of pathogenic microorganism detection, and the DETECTR system is based on Cas12a, and once the target DNA-specific sgRNA is detected, the Cas12a’s endonuclease activity would be activated and cleave both the target and non-target genes. In order to verify if there are related parts, we searched the iGEM Biological Parts library and picked BBa_K3136004. This is a biologic part submitted by iGEM19_Shanghai_HS_United in 2019, and the team provided a complete reaction system to detect the presence of Listeria monocytogenes. Our team developed a similar reaction platform to detect pathogenic microorganisms, such as Helicobacter Pylori, salmonella, and shigella, adding data from in vitro DETECTR reaction system.
What’s more, once the endonuclease activity of FnCas12a was activated, this protein could also cut the non-target DNA fragments. Based on this phenomenon, we employed DNA probes with the 6-FAM flag on its 5’-terminal. When there was sgRNAs of target genes were recognized by FnCas12a, the probes would also be cleaved, then we could detect the fluorescence.
We synthesized four DNA fragments amplified from the pathogenic microorganisms we chose, and insert them in the pUC57 vector. Next, we purified the FnCas12a protein and synthesized probes with the 6-FAM flag. Then, we mixed those materials and the reaction buffer to develop the in vitro reaction system, and we also changed the concentration of the target genes. Finally, we detected the fluorescence of the reaction system and measured the activity of FnCas12a (Figure 1).
Engineering Success
FnCas12a, which is amplified from Francisella novicida, is a new class II family of CRISPR-Cas RNA-programmable endonucleases with unique features that make it a very attractive alternative or complement to Cas9 for genome engineering.
a) Construction of pathogenic micro-organisms expression plasmids We designed 5 plasmids: the FnCas12 protein expression plasmid, 16S, cagA, ipaH, and invA expression plasmids. Among them, the DNA fragments 16S and cagA are amplified from the genome of Helicobacter Pylori, and the gene fragments ipaH and invA are amplified from salmonella and shigella genomic DNA respectively.
In order to construct our plasmids, we let the company synthesize the DNA fragments, FnCas12 was inserted into the pET28a vector, and the fragments 16S, cagA, ipaH, and invA were inserted into the pUC57 vector. The constructed plasmids were contained in E. coli strains, we streak inoculated them on LB solid medium plates containing corresponding antibiotics, and incubate them at 37℃ overnight.
b) Verification of the microorganisms expression plasmids
We used TAE agarose gel electrophoresis to testify the presence of oligo DNA in the plasmid by performing PCR and then doing gel electrophoresis of the amplicons (Figure 2).
Our results show that a band of 200bp to 400bp is present in cagA, 16S, invA, and ipaH, but not in negative control (NC) lanes. Because oligo DNA has a size between 200bp to 400bp, our result supports the fact that the plasmids contain desired oligo DNA. The four plasmid transformations were successful.
Expression and purification of Cas12a protein
We transformed the pET28a-FnCas12a expression plasmid into E. coli BL21(DE3) competent cells, and cultured at 37℃ overnight (Figure 1A). we inoculated a single colony into LB (Kana+) culture medium, incubated overnight, and then transferred the cultured medium into 1L fresh LB (Kana+) culture medium. We induced the expression of FnCas12a with IPTG when the OD600 was around 0.6-1.0, and cultured at 16℃ for 12h. Subsequently, we used nickel affinity purification to purify the acquired Cas12a proteins from other proteins in E. coli (Figure 1B).
A. Incubate the plasmid pET28a-FnCas12a containing BL21(DE3). B. SDS-PAGE electrophoresis gel of Cas12a protein compared to nonspecific protein impurities.
Bicinchoninic Acid Assay (BCA)
Cas12a protein has a size of 130kDa. The SDS-PAGE electrophoresis result indicates that the Cas12a protein is present in the solution we collected at 130kDa, and not present in the nonspecific protein impurities. Thus, Cas12a proteins were expressed and purified with high quality.
Then, we tested the concentration of Cas12a protein by Bicinchoninic Acid Assay (BCA), using SpectraMax i3x Multi-Mode Microplate Reader with the absorption peak at 562nm (Figure 2).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1591
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 365
Illegal BglII site found at 1544
Illegal BglII site found at 1578
Illegal BglII site found at 1623
Illegal BglII site found at 2402
Illegal BglII site found at 3383 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 3184
Illegal AgeI site found at 3253 - 1000COMPATIBLE WITH RFC[1000]