Difference between revisions of "Part:BBa K4150003"
Jacky CHIN (Talk | contribs) |
Jacky CHIN (Talk | contribs) |
||
| Line 3: | Line 3: | ||
<partinfo>BBa_K4150003 short</partinfo> | <partinfo>BBa_K4150003 short</partinfo> | ||
| − | <span style="color:#00000000">This</span> The GFP sequence is designed based on gfpmut1 ([[Part:BBa_K823039]]) connected with a His tag and a GS linker at N-terminus. | + | <span style="color:#00000000">This</span> The GFP sequence is designed based on gfpmut1 ([[Part:BBa_K823039]]) connected with a His tag and a GS linker at N-terminus. The part was used as reporter gene to demonstrate the funcationality of the engineered T7 bacteriophage. |
| + | |||
| + | ===Characterization=== | ||
| + | |||
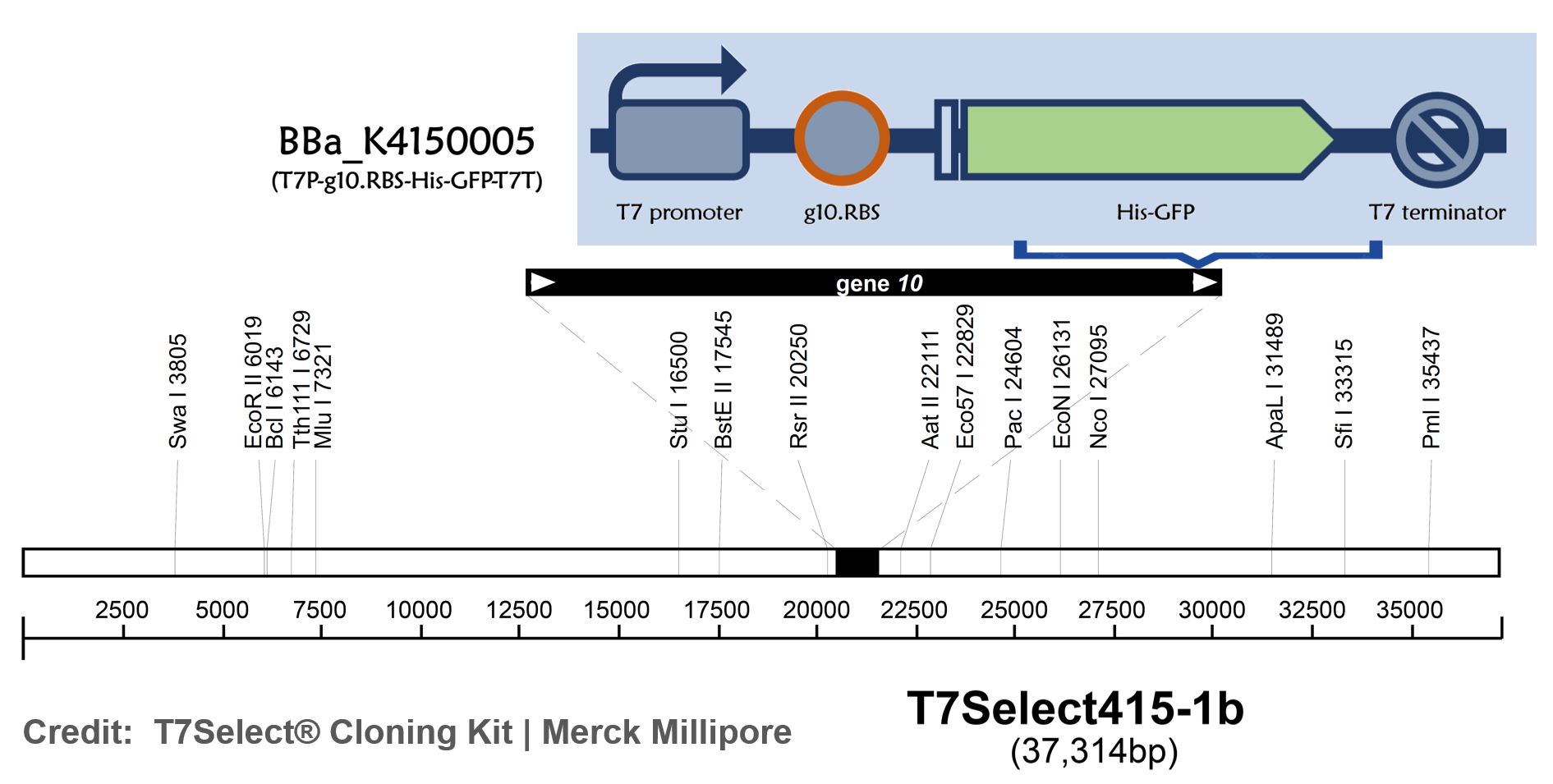
| + | <span style="color:#00000000">This</span>T7 bacteriophage genome was edited with the BioBrick of T7P-g10.RBS-His-Ova-T7T ([[Part:BBa K4150006]])in the end of T7 gene 10 using the T7Select® 415-1 Cloning Kit (Merck Millipore) according to the manufacturer’s instruction. | ||
| + | |||
| + | <br> | ||
| + | [[File:T--Mingdao--2022--5-3.jpeg|700px|center]] | ||
| + | <br> | ||
| + | |||
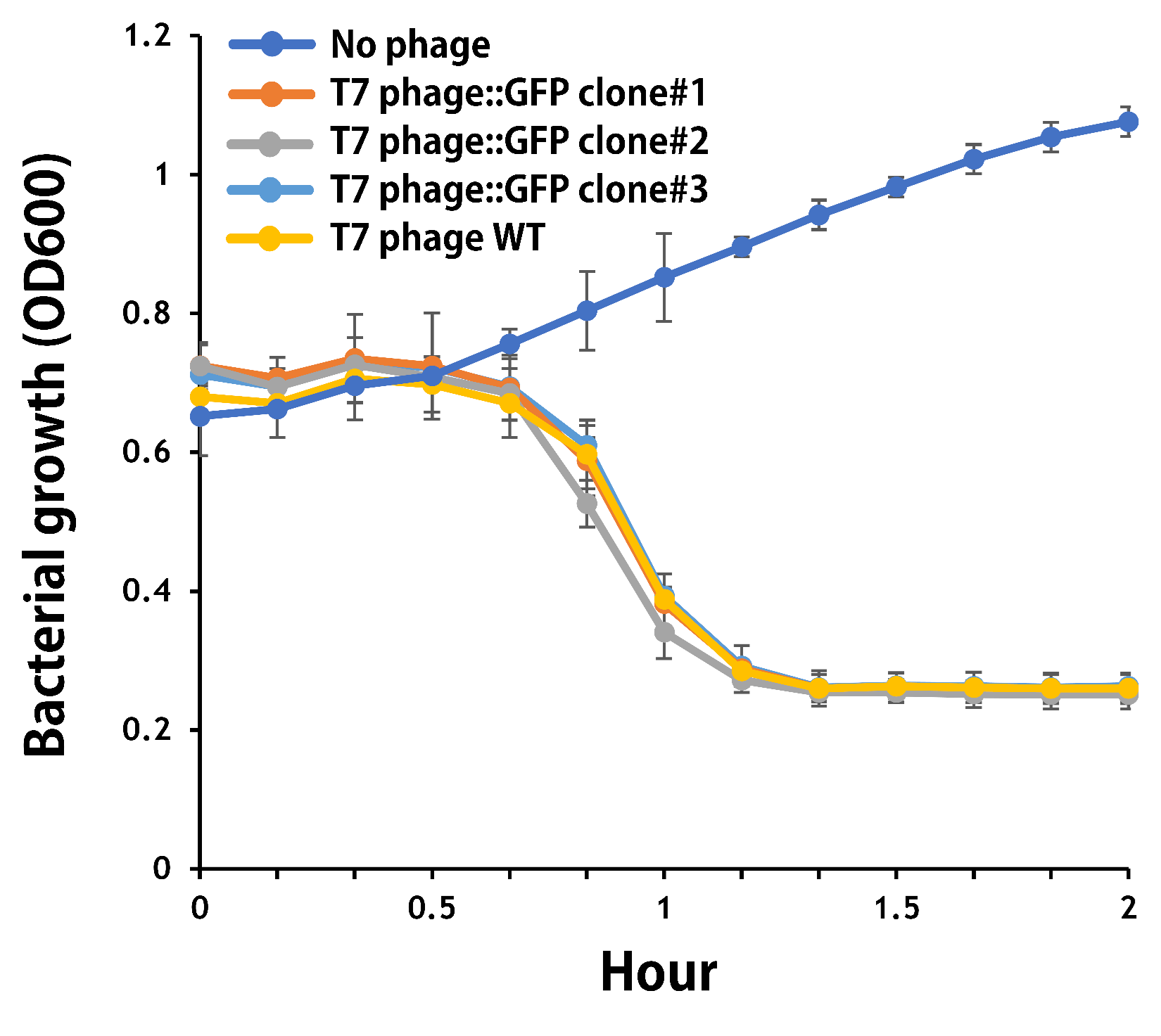
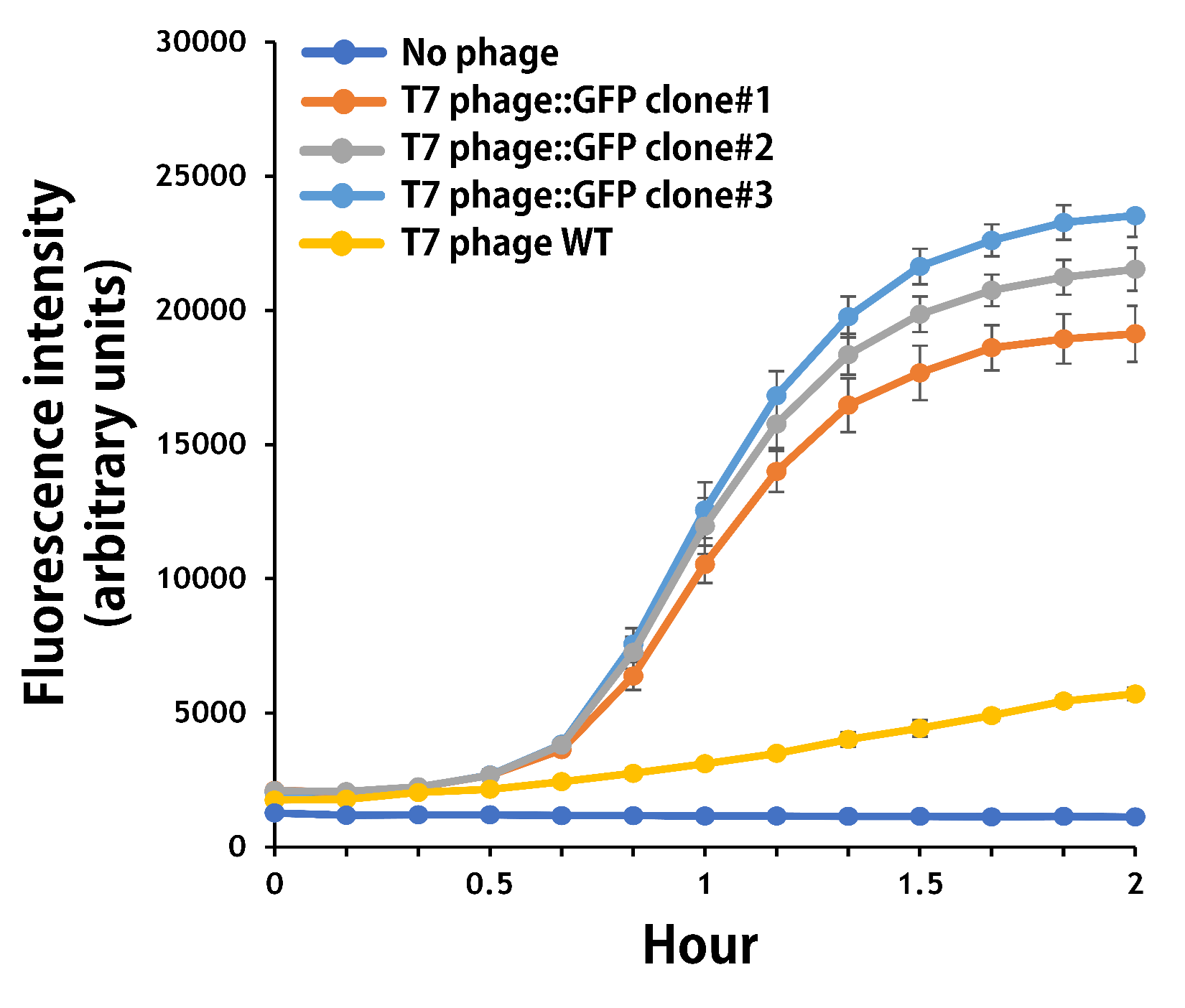
| + | <span style="color:#00000000">This</span>To test the functionality of T7 phage::T7P-g10.RBS-His-GFP-T7T, the mouse intestinal commensal E. coli isolates were grown to an OD around 0.6 in the M9 minimal medium supplemented with casamino acids (0.2%), D-glucose (0.3%), vitamin B1 (1 mg/ml), MgSO4 (0.2 mM), CaCl2 (0.1 mM)<ref>Vinay M, Franche N, Grégori G, Fantino JR, Pouillot F, Ansaldi M. Phage-Based Fluorescent Biosensor Prototypes to Specifically Detect Enteric Bacteria Such as E. coli and Salmonella enterica Typhimurium. PLoS One. 2015 Jul 17;10(7):e0131466. doi: 10.1371/journal.pone.0131466. PMID: 26186207; PMCID: PMC4506075.</ref>, followed by infected with the engineered phages at a MOI of 5 for 2 hr at 37°C. The data were read for OD600 and GFP at an ex/em = 483/513 nm every 10 min. Compared to wild-type T7 phage infection, the engineered phage::T7P-g10.RBS-His-GFP-T7T can infect and kill the E. coli in a similar way (Fig. 3). In addition, the engineered phages are able to transfer the GFP reporter cassette into E. coli and make GFP production at the beginning of 40 min (Fig. 4), which is consistent with the time starting to lyse the bacteria (Fig. 3). | ||
| + | |||
| + | <br> | ||
| + | [[File:T--Mingdao--2022--6-12.png|500px|left]] | ||
| + | |||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | <b>Figure 3 |</b> The infection of the wild-type and the engineered T7 phages on a mouse intestinal commensal E. coli. The E. coli were growing to an OD around 0.6 in the wells of a 96-well plate (Falcon® 96-well Black/Clear Flat Bottom) at 37°C with 200 μl of the supplemented M9 media, followed by the phage (as indicated) infection at a MOI of 5. The values of OD600 were read every 10 min till 2 hr by a microplate reader (Synergy H1 Hybrid Multi-Mode Reader - BioTek Instruments). The growth of E. coli without phages was set as a control. | ||
| + | |||
| + | <br> | ||
| + | [[File:T--Mingdao--2022--6-13.png|500px|left]] | ||
| + | |||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | <b>Figure 4 |</b> The production of GFP by the engineered T7 phage::T7P-g10.RBS-His-GFP-T7T. The E. coli were growing to an OD around 0.6 in the wells of a 96-well plate (Falcon® 96-well Black/Clear Flat Bottom) at 37°C with 200μl of the supplemented M9 media, followed by the phage (as indicated) infection at a MOI of 5. The values of fluorescence intensity were read at an ex/em = 483/513 nm every 10 min till 2 hr by a microplate reader (Synergy H1 Hybrid Multi-Mode Reader - BioTek Instruments). The background levels were measured with the controls of E. coli without phages or infected with wild-type phages. | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 07:08, 20 September 2022
His-GFP
This The GFP sequence is designed based on gfpmut1 (Part:BBa_K823039) connected with a His tag and a GS linker at N-terminus. The part was used as reporter gene to demonstrate the funcationality of the engineered T7 bacteriophage.
Characterization
ThisT7 bacteriophage genome was edited with the BioBrick of T7P-g10.RBS-His-Ova-T7T (Part:BBa K4150006)in the end of T7 gene 10 using the T7Select® 415-1 Cloning Kit (Merck Millipore) according to the manufacturer’s instruction.
ThisTo test the functionality of T7 phage::T7P-g10.RBS-His-GFP-T7T, the mouse intestinal commensal E. coli isolates were grown to an OD around 0.6 in the M9 minimal medium supplemented with casamino acids (0.2%), D-glucose (0.3%), vitamin B1 (1 mg/ml), MgSO4 (0.2 mM), CaCl2 (0.1 mM)[1], followed by infected with the engineered phages at a MOI of 5 for 2 hr at 37°C. The data were read for OD600 and GFP at an ex/em = 483/513 nm every 10 min. Compared to wild-type T7 phage infection, the engineered phage::T7P-g10.RBS-His-GFP-T7T can infect and kill the E. coli in a similar way (Fig. 3). In addition, the engineered phages are able to transfer the GFP reporter cassette into E. coli and make GFP production at the beginning of 40 min (Fig. 4), which is consistent with the time starting to lyse the bacteria (Fig. 3).
Figure 3 | The infection of the wild-type and the engineered T7 phages on a mouse intestinal commensal E. coli. The E. coli were growing to an OD around 0.6 in the wells of a 96-well plate (Falcon® 96-well Black/Clear Flat Bottom) at 37°C with 200 μl of the supplemented M9 media, followed by the phage (as indicated) infection at a MOI of 5. The values of OD600 were read every 10 min till 2 hr by a microplate reader (Synergy H1 Hybrid Multi-Mode Reader - BioTek Instruments). The growth of E. coli without phages was set as a control.
Figure 4 | The production of GFP by the engineered T7 phage::T7P-g10.RBS-His-GFP-T7T. The E. coli were growing to an OD around 0.6 in the wells of a 96-well plate (Falcon® 96-well Black/Clear Flat Bottom) at 37°C with 200μl of the supplemented M9 media, followed by the phage (as indicated) infection at a MOI of 5. The values of fluorescence intensity were read at an ex/em = 483/513 nm every 10 min till 2 hr by a microplate reader (Synergy H1 Hybrid Multi-Mode Reader - BioTek Instruments). The background levels were measured with the controls of E. coli without phages or infected with wild-type phages.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 692
- ↑ Vinay M, Franche N, Grégori G, Fantino JR, Pouillot F, Ansaldi M. Phage-Based Fluorescent Biosensor Prototypes to Specifically Detect Enteric Bacteria Such as E. coli and Salmonella enterica Typhimurium. PLoS One. 2015 Jul 17;10(7):e0131466. doi: 10.1371/journal.pone.0131466. PMID: 26186207; PMCID: PMC4506075.