Difference between revisions of "Part:BBa K3725022"
| Line 5: | Line 5: | ||
<b>Overview</b> | <b>Overview</b> | ||
| − | The Improved Fusarium Toehold w/ GFP Reporter part, is designed to be used in conjunction with the T7 F. oxysporum f.sp. lycopersici trigger (Part BBa_K3725070) in order to express GFP as a part of the engineered toehold switch system. The described structure was designed using NUPACK (Nucleic Acid Package) software’s Design feature. This was done by finding a unique gene of the F. oxysporum species that encodes for pathogenicity: the FRP1 gene. After inputting the FRP1 gene sequence into the NUPACK software using an input code given by Takashi et. al., pairs of trigger sequences and switch sequences were outputted. The trigger sequences given were 36 base-pair long sequences from the FRP1 gene, and the switch sequences given were reverse complementary to the trigger sequences. The pairs were ordered by normalized ensemble defect. The Improved Fusarium Toehold w/ GFP reporter was the generated pair with the second-lowest normalized ensemble defect. We ordered the insert in a pUCIDT Amp plasmid from Integrated DNA Technologies. | + | The Improved Fusarium Toehold w/ GFP Reporter part, is designed to be used in conjunction with the T7 <i>F. oxysporum f.sp. lycopersici</i> trigger (Part BBa_K3725070) in order to express GFP as a part of the engineered toehold switch system. The described structure was designed using NUPACK (Nucleic Acid Package) software’s Design feature. This was done by finding a unique gene of the <i>F. oxysporum</i> species that encodes for pathogenicity: the FRP1 gene. After inputting the FRP1 gene sequence into the NUPACK software using an input code given by Takashi et. al., pairs of trigger sequences and switch sequences were outputted. The trigger sequences given were 36 base-pair long sequences from the FRP1 gene, and the switch sequences given were reverse complementary to the trigger sequences. The pairs were ordered by normalized ensemble defect. The Improved Fusarium Toehold w/ GFP reporter was the generated pair with the second-lowest normalized ensemble defect. We ordered the insert in a pUCIDT Amp plasmid from Integrated DNA Technologies. |
<strong>Description</strong> | <strong>Description</strong> | ||
| Line 16: | Line 16: | ||
<b>Design</b> | <b>Design</b> | ||
| − | The construction of a disease-specific biosensor required us to find a gene unique to the pathogen. When the switch turns on and GFP is expressed, we can confirm that the specific pathogen is present. For the detection of Fusarium oxysporum f. sp. lycopersici, Lambert iGEM focused on the FRP1 gene. This gene was selected because it was required for pathogenicity and was unique to the species of interest. Biosafety note, the trigger sequence is not the full transcript sequence and therefore poses limited biosafety. We obtained the sequence via UniProt, an online database of protein sequences. Lambert iGEM used the code from Takahashi et. al provided by Megan McSweeney from the Styczynski Lab at the Georgia Institute of Technology to design the switch and trigger sequences on NUPACK. The team selected the pair from NUPACK with the lowest normalized ensemble defect (NED) to maximize the chances of successful compatibility. Once we obtained the sequences for the toehold pair, we constructed the toehold and trigger via SnapGene. | + | The construction of a disease-specific biosensor required us to find a gene unique to the pathogen. When the switch turns on and GFP is expressed, we can confirm that the specific pathogen is present. For the detection of <i>Fusarium oxysporum f. sp. lycopersici</i>, Lambert iGEM focused on the FRP1 gene. This gene was selected because it was required for pathogenicity and was unique to the species of interest. Biosafety note, the trigger sequence is not the full transcript sequence and therefore poses limited biosafety. We obtained the sequence via UniProt, an online database of protein sequences. Lambert iGEM used the code from Takahashi et. al provided by Megan McSweeney from the Styczynski Lab at the Georgia Institute of Technology to design the switch and trigger sequences on NUPACK. The team selected the pair from NUPACK with the lowest normalized ensemble defect (NED) to maximize the chances of successful compatibility. Once we obtained the sequences for the toehold pair, we constructed the toehold and trigger via SnapGene. |
This part is different from Part BBa_K3725020, the Fusarium Toehold w/ GFP reporter, in that there are differences in the sequence that change the hairpin loop structure of the toehold switch. This was the pair with the second-lowest normalized ensemble defect from NUPACK. As compared to the Fusarium Toehold w/ GFP reporter, the free energy of the hairpin loop of our redesigned part is higher, at -25.30 kcal/mol versus -26.10 kcal/mol. | This part is different from Part BBa_K3725020, the Fusarium Toehold w/ GFP reporter, in that there are differences in the sequence that change the hairpin loop structure of the toehold switch. This was the pair with the second-lowest normalized ensemble defect from NUPACK. As compared to the Fusarium Toehold w/ GFP reporter, the free energy of the hairpin loop of our redesigned part is higher, at -25.30 kcal/mol versus -26.10 kcal/mol. | ||
Latest revision as of 22:43, 21 October 2021
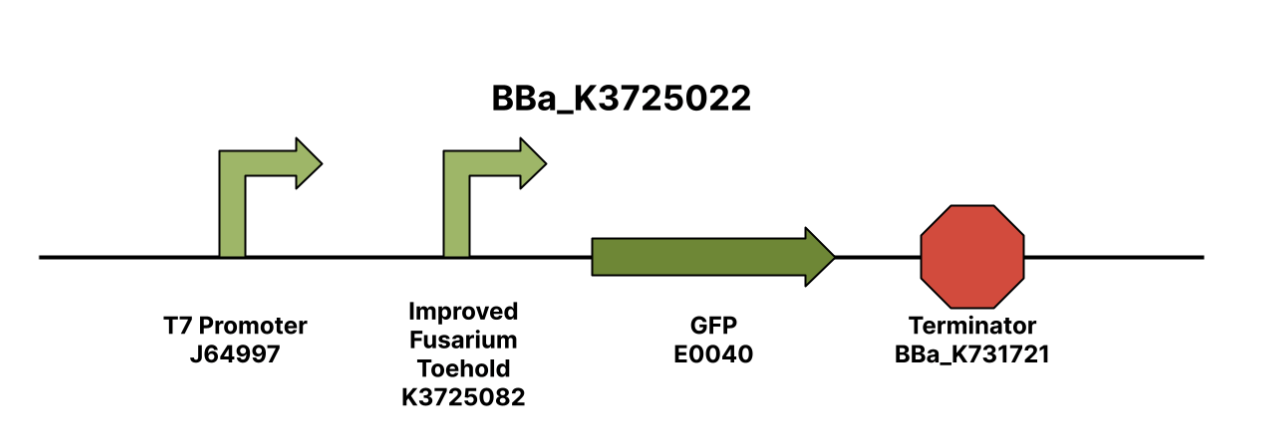
Improved Fusarium Toehold w/ GFP Reporter
Overview
The Improved Fusarium Toehold w/ GFP Reporter part, is designed to be used in conjunction with the T7 F. oxysporum f.sp. lycopersici trigger (Part BBa_K3725070) in order to express GFP as a part of the engineered toehold switch system. The described structure was designed using NUPACK (Nucleic Acid Package) software’s Design feature. This was done by finding a unique gene of the F. oxysporum species that encodes for pathogenicity: the FRP1 gene. After inputting the FRP1 gene sequence into the NUPACK software using an input code given by Takashi et. al., pairs of trigger sequences and switch sequences were outputted. The trigger sequences given were 36 base-pair long sequences from the FRP1 gene, and the switch sequences given were reverse complementary to the trigger sequences. The pairs were ordered by normalized ensemble defect. The Improved Fusarium Toehold w/ GFP reporter was the generated pair with the second-lowest normalized ensemble defect. We ordered the insert in a pUCIDT Amp plasmid from Integrated DNA Technologies.
Description
Toehold biosensors, which are composed of a switch and trigger, are highly orthogonal riboregulators that activate translation in response to a specific RNA sequence. The switch is composed of a hairpin loop structure that represses translation through its complementary bases in between the ribosomal binding site and the start codon, which is followed by a linker sequence. Once the toehold is exposed to the trigger sequence, the complementary base pairs on the trigger will bind to the toehold, which exposes the ribosomal binding site. RNA polymerase can then bind to the RBS and initiate translation of the reporter protein.
Design
The construction of a disease-specific biosensor required us to find a gene unique to the pathogen. When the switch turns on and GFP is expressed, we can confirm that the specific pathogen is present. For the detection of Fusarium oxysporum f. sp. lycopersici, Lambert iGEM focused on the FRP1 gene. This gene was selected because it was required for pathogenicity and was unique to the species of interest. Biosafety note, the trigger sequence is not the full transcript sequence and therefore poses limited biosafety. We obtained the sequence via UniProt, an online database of protein sequences. Lambert iGEM used the code from Takahashi et. al provided by Megan McSweeney from the Styczynski Lab at the Georgia Institute of Technology to design the switch and trigger sequences on NUPACK. The team selected the pair from NUPACK with the lowest normalized ensemble defect (NED) to maximize the chances of successful compatibility. Once we obtained the sequences for the toehold pair, we constructed the toehold and trigger via SnapGene.
This part is different from Part BBa_K3725020, the Fusarium Toehold w/ GFP reporter, in that there are differences in the sequence that change the hairpin loop structure of the toehold switch. This was the pair with the second-lowest normalized ensemble defect from NUPACK. As compared to the Fusarium Toehold w/ GFP reporter, the free energy of the hairpin loop of our redesigned part is higher, at -25.30 kcal/mol versus -26.10 kcal/mol.
Experience
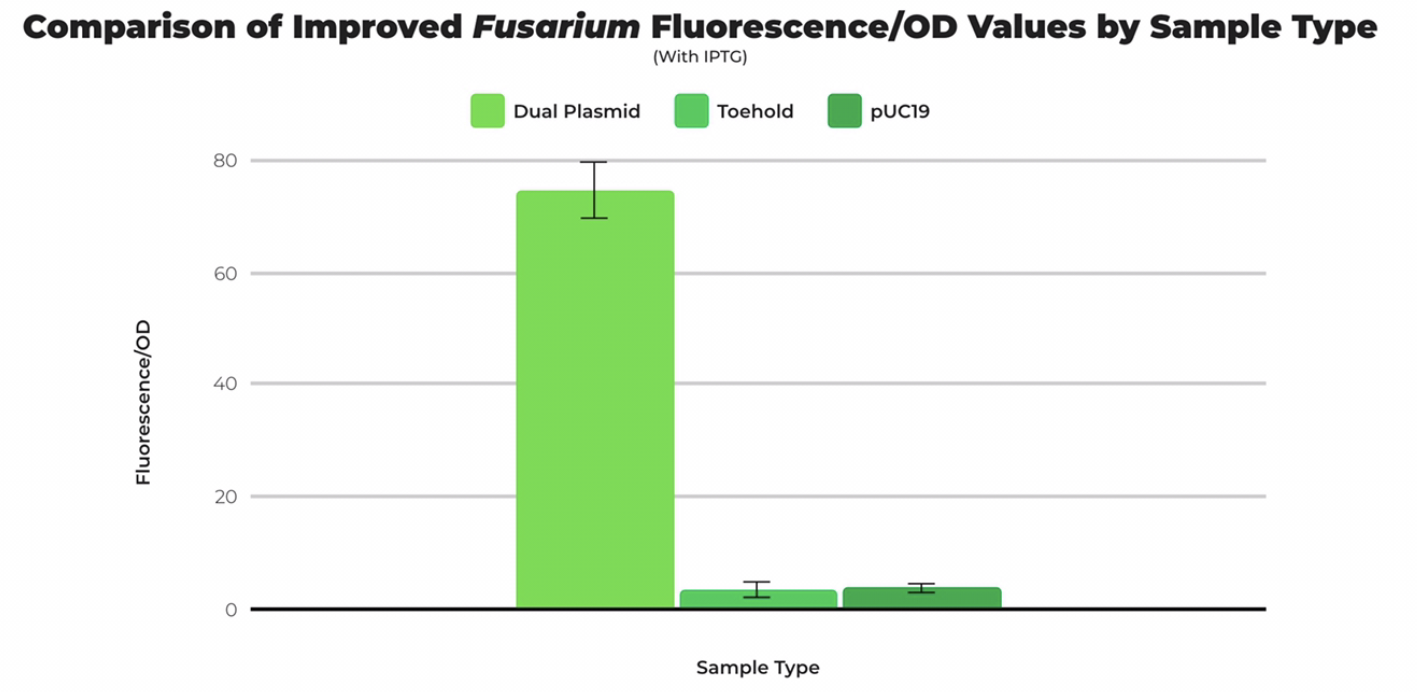
To test the compatibility of the Fusarium trigger sequence (Part BBa_K3725070) with the improved toehold sequence, a dual-plasmid transformation was performed using electrocompetent cells. After growing liquid cultures, running a miniprep, and performing a restriction digest, we confirmed dual plasmid success by seeing two bands. Fluorescence was measured in comparison with cells transformed with only the toehold part and pUC19 (positive control). All samples were divided by optical density to obtain a standardized unit. The fluorescence per optical density unit measurement of cells in the dual plasmid transformation was significantly higher than cells with toehold only and pUC19, confirming that the toehold-trigger pair is compatible.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 861
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 777