Difference between revisions of "Part:BBa K2927006"
Alice-Huang (Talk | contribs) |
Alice-Huang (Talk | contribs) |
||
| Line 20: | Line 20: | ||
<div style='text-align: center;'> | <div style='text-align: center;'> | ||
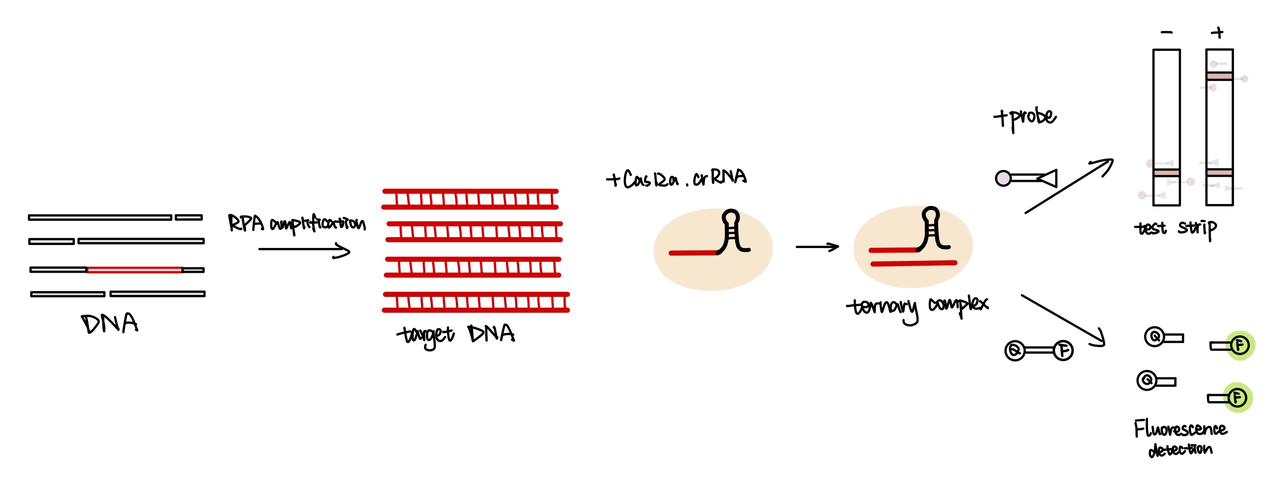
| − | [[File:T--GreatBay SZ--cas12a | + | [[File:T--GreatBay SZ--cas12a detection.png|600px|thumb|center]] |
| − | <b> 【fig.1】Cas12a | + | <b> 【fig.1】Cas12a detection </b> |
| + | |||
| + | </div> | ||
<div style='text-align: center;'> | <div style='text-align: center;'> | ||
| Line 41: | Line 43: | ||
[[File:T--GreatBay_SZ--cas12a_orthogonal_experiment_heat_map.png|400px|thumb|center]] | [[File:T--GreatBay_SZ--cas12a_orthogonal_experiment_heat_map.png|400px|thumb|center]] | ||
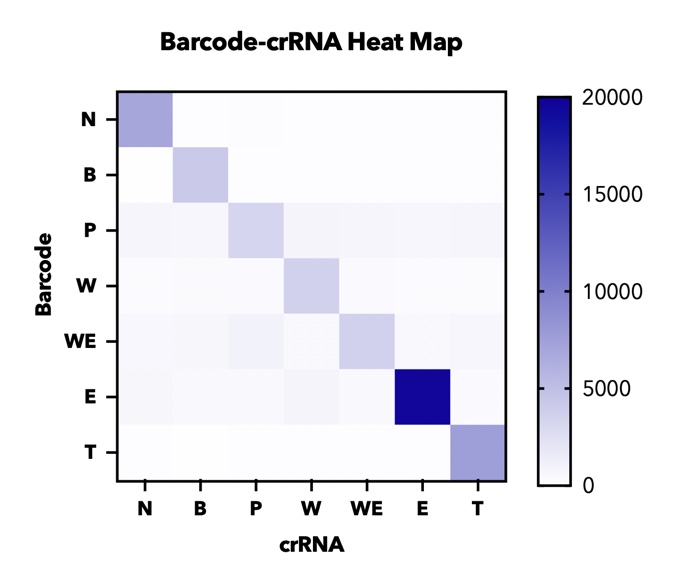
| − | <b> 【fig. | + | <b> 【fig.4】Heatmap of endpoint fluorescence values from in cas12a fluorescence detection of all combinations of 7 barcodes and 7 crRNAs assessing specificity of each barcode-crRNA pair. </b> |
</div> | </div> | ||
Revision as of 18:19, 21 October 2021
guide RNA of LbCas12a
The CRISPR/Case system uses guide RNA (gRNA) and crRNAs to recognize their targets. The gRNA sequence binds to Cas12a protein, which promotes crRNA to recognize its target double-stranded DNA through sequence complementation.
Characterization by iGEM21_GreatBay_SZ
- Group: iGEM21_GreatBay_SZ
- Author: TianYi Huang
We attached the repeat sequence to guide RNA used to recognize their targets(Part:BBa_K3859013). The crRNA will bind with cas12a enzyme, then the target DNA will be recobgnised and unwound by cas12a. After target DNA binds with guide RNA, cas12a will be activated and cleveage the sequence around it. In our project, we primarily used Cas12a detection technology for the identification of spores. This decision was made with reference to the methodology used in the HOLMES system
Experienment and result from iGEM21_GreatBay_SZ
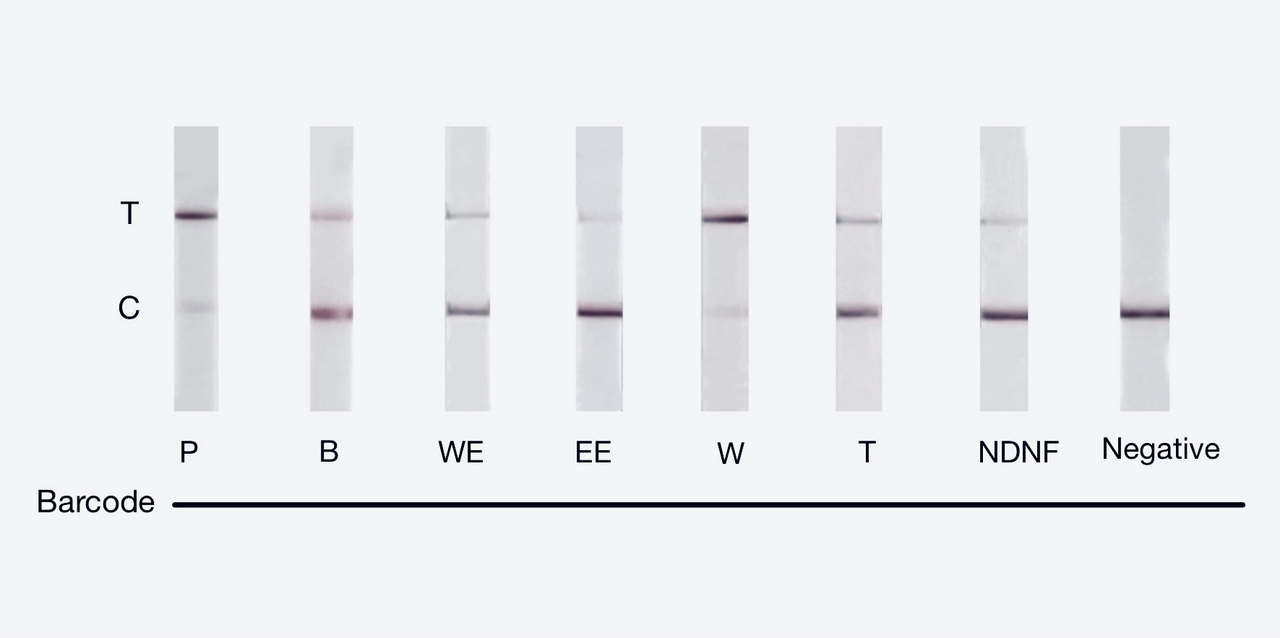
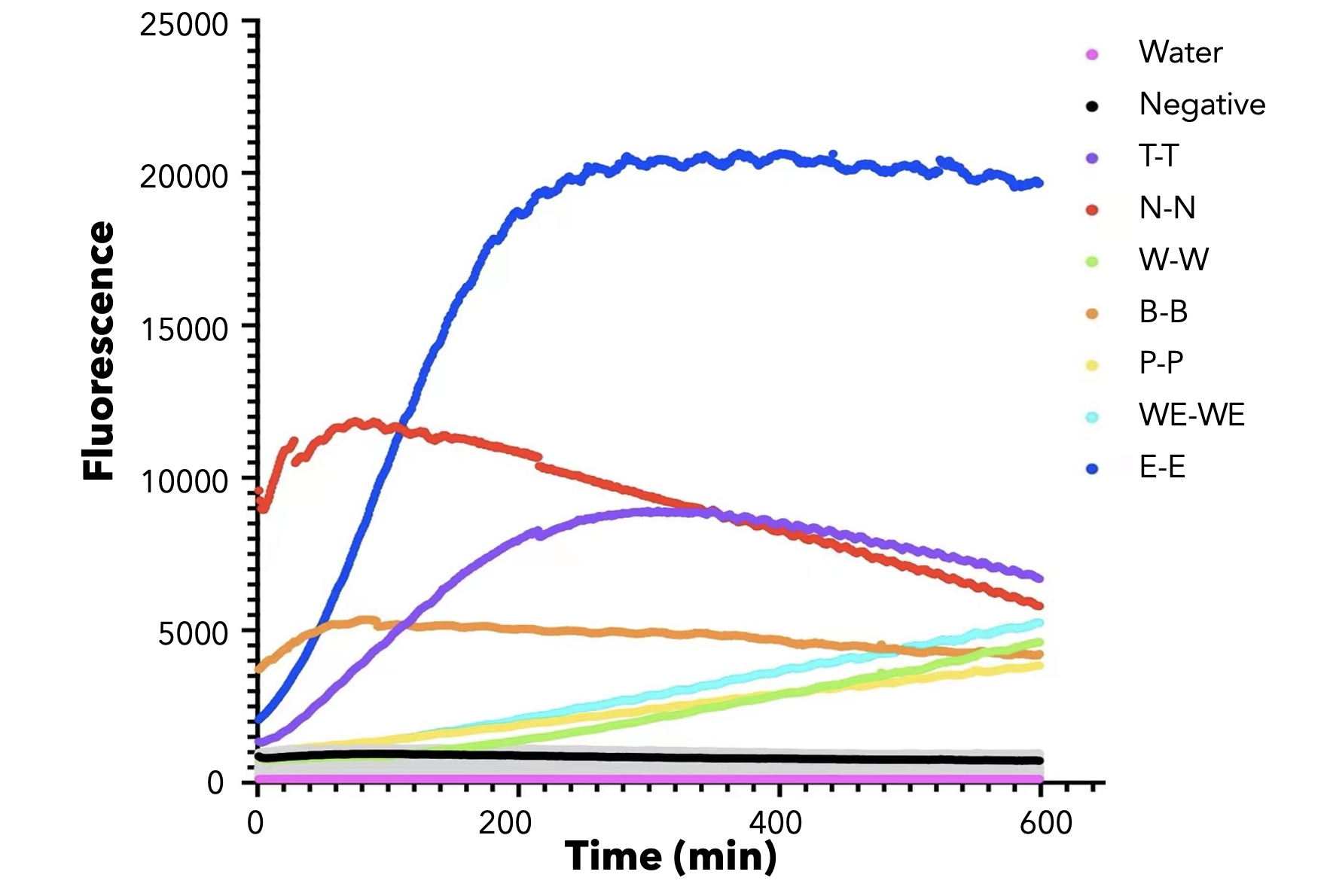
We used both the test strip and the florencence method in vitro. For the test strip, we successfully got the correct result for all the barcodes(fig.2). For the florencence method, we got the graphs of the florencence against time(fig.3)[2], which were proofed right by our model.
In addition, in order to test the specificity of our barcode design. We constructed 7 barcodes and their matching CRISPR RNAs (crRNAs) and assayed all permutations in vitro using cas12a florencence detection(fig.4).
【fig.2】Cas12a test strip result
【fig.3】Cas12a fluorescence detection result. On the left of the symbol "-" refers to barcode name; On the right of the symbol "-" refers to crRNA name. All results for barcode mismatch with crRNA are shown in grey. E.g. "T-T" means the mixture of T-barcode and T-crRNA.
In addition, in order to test the specificity of our barcode design. We constructed 7 barcodes and their matching CRISPR RNAs (crRNAs) and assayed all permutations in vitro using cas12a florencence detection(fig.3)[2].
【fig.4】Heatmap of endpoint fluorescence values from in cas12a fluorescence detection of all combinations of 7 barcodes and 7 crRNAs assessing specificity of each barcode-crRNA pair.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]