Difference between revisions of "Part:BBa K3863006"
Alisaleong (Talk | contribs) |
Alisaleong (Talk | contribs) |
||
| Line 14: | Line 14: | ||
<partinfo>BBa_K3863006 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3863006 SequenceAndFeatures</partinfo> | ||
| − | < | + | ==Team PuiChing_Macau 2021: Break down monosaccharides== |
| + | ===Validation of construct expression using RT-qPCR=== | ||
| + | https://2021.igem.org/wiki/images/7/7e/T--PuiChing_Macau--1aa.jpg | ||
| + | <p>1a) Amylase (BBa_K3863006) qPCR</p> | ||
| + | |||
| + | https://2021.igem.org/wiki/images/9/92/T--PuiChing_Macau--1b.jpg | ||
| + | <p>1b) Glucoamylase (BBa_K3863006) qPCR</p> | ||
| + | |||
| + | <p> | ||
| + | To future confirm that our constructs have the expression level, we have performed RT-qPCR | ||
| + | The RT-qPCR results show that we have successfully assembled target genes K3863006 to the vector (pETDuet). | ||
| + | </p> | ||
| + | |||
| + | ==Reference== | ||
<p>[1]Masayoshi Sakaguchi,corresponding author1 Yudai Matsushima,1 Toshiyuki Nankumo,1 Junichi Seino,1 Satoshi Miyakawa,1 Shotaro Honda,1 Yasusato Sugahara,1 Fumitaka Oyama,1 and Masao Kawakita1,2, (2014) (Glucoamylase of Caulobacter crescentus CB15: cloning and expression in Escherichia coli and functional identification)</p> | <p>[1]Masayoshi Sakaguchi,corresponding author1 Yudai Matsushima,1 Toshiyuki Nankumo,1 Junichi Seino,1 Satoshi Miyakawa,1 Shotaro Honda,1 Yasusato Sugahara,1 Fumitaka Oyama,1 and Masao Kawakita1,2, (2014) (Glucoamylase of Caulobacter crescentus CB15: cloning and expression in Escherichia coli and functional identification)</p> | ||
Revision as of 15:30, 21 October 2021
a-amylase-HA-glucoamylase-his
This is a composite part designed to break down starch in food waste into monosaccharides. Amylase: E. coli periplasmic α-amylase gene malS. The protein sequence in this part is based on K523001 from Edinburgh 2011. Glucoamylase: Caulobacter crescentus CB15, caulobacter vibrioides gene for glucoamylase(GenBank: AB813000)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1287
Illegal NotI site found at 2146 - 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 3884
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 2441
Illegal NgoMIV site found at 2633
Illegal NgoMIV site found at 2637
Illegal NgoMIV site found at 3275
Illegal NgoMIV site found at 3649
Illegal NgoMIV site found at 3803
Illegal NgoMIV site found at 3824
Illegal AgeI site found at 423
Illegal AgeI site found at 1588
Illegal AgeI site found at 2393 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 3026
Team PuiChing_Macau 2021: Break down monosaccharides
Validation of construct expression using RT-qPCR
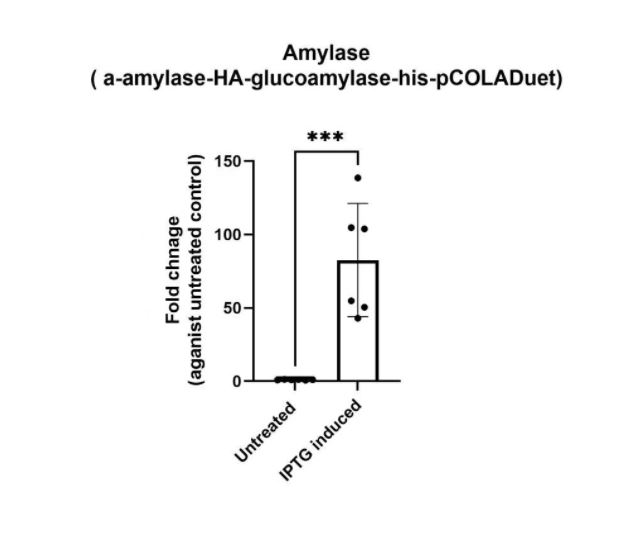
1a) Amylase (BBa_K3863006) qPCR
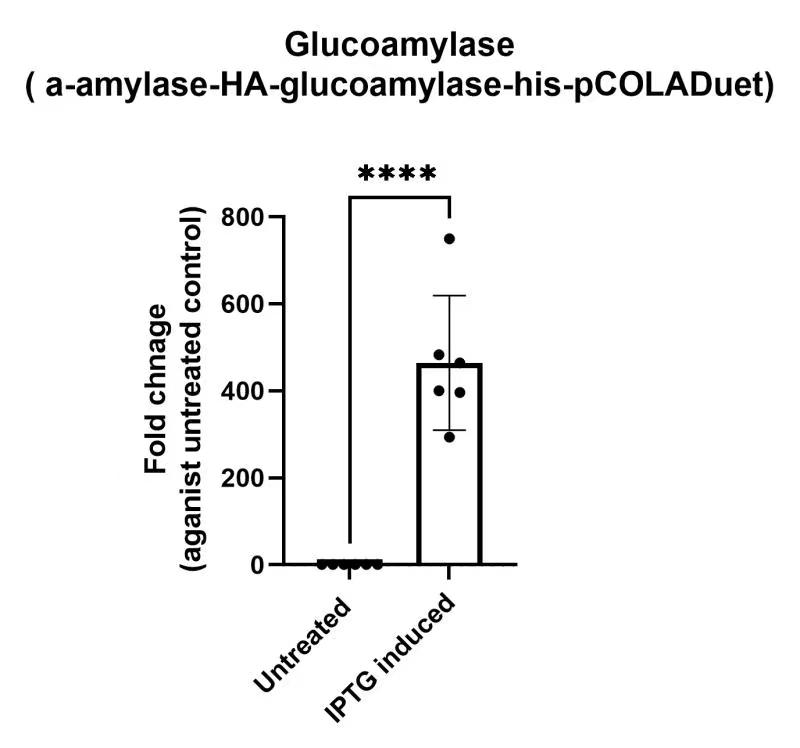
1b) Glucoamylase (BBa_K3863006) qPCR
To future confirm that our constructs have the expression level, we have performed RT-qPCR The RT-qPCR results show that we have successfully assembled target genes K3863006 to the vector (pETDuet).
Reference
[1]Masayoshi Sakaguchi,corresponding author1 Yudai Matsushima,1 Toshiyuki Nankumo,1 Junichi Seino,1 Satoshi Miyakawa,1 Shotaro Honda,1 Yasusato Sugahara,1 Fumitaka Oyama,1 and Masao Kawakita1,2, (2014) (Glucoamylase of Caulobacter crescentus CB15: cloning and expression in Escherichia coli and functional identification)
