Difference between revisions of "Part:BBa K3747301"
| Line 24: | Line 24: | ||
https://2021.igem.org/wiki/images/5/5d/T--Wageningen_UR--wetlab_MKS_HP.png | https://2021.igem.org/wiki/images/5/5d/T--Wageningen_UR--wetlab_MKS_HP.png | ||
| − | |||
==References== | ==References== | ||
1. Y. Chen, J. M. L. Ho, D. L. Shis, et al., Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 9, 1–8 (2018). | 1. Y. Chen, J. M. L. Ho, D. L. Shis, et al., Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 9, 1–8 (2018). | ||
Latest revision as of 13:19, 21 October 2021
Plux/lac_dE
Plux/lac_dE hybrid promotor
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 583
- 12INCOMPATIBLE WITH RFC[12]Illegal SpeI site found at 583
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 583
- 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 583
- 1000COMPATIBLE WITH RFC[1000]
By combining multiple operator sites in a promoter region, a “hybrid promoter” can be created that responds to multiple input signals. This creates a logic transcriptional AND gate, of which the dynamic range can be further tuned by changing -35 and -10 sites. This E. coli promoter, Plux/lac, was developed by Chen et al. (2018) [1] and contains operator sites for LacI repression and LuxR activation. Transcription is fully activated when LacI is absent and LuxR is present. Different -10 sites “E” and “F” influence the transcription rate when only one of the two input signals is present (Figure 1).
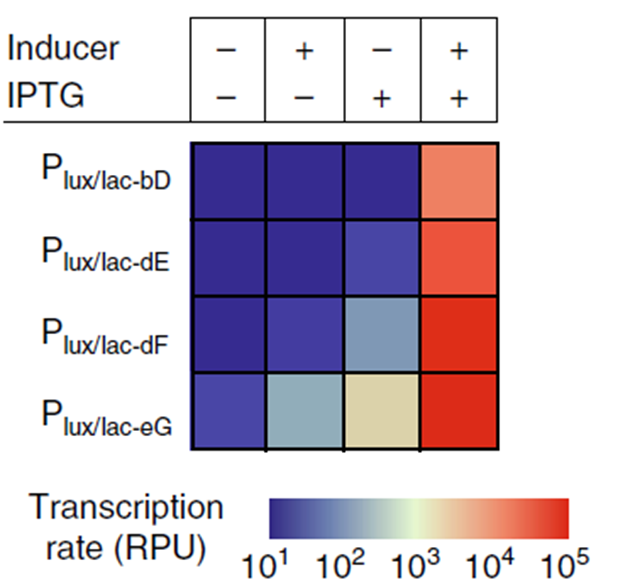
References
1. Y. Chen, J. M. L. Ho, D. L. Shis, et al., Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 9, 1–8 (2018).
