Difference between revisions of "Part:BBa K3747301"
| Line 23: | Line 23: | ||
https://2021.igem.org/wiki/images/5/5d/T--Wageningen_UR--wetlab_MKS_HP.png | https://2021.igem.org/wiki/images/5/5d/T--Wageningen_UR--wetlab_MKS_HP.png | ||
| + | |||
| + | <a href="https://2021.igem.org/wiki/index.php?title=Team:Wageningen" | ||
| + | >BB2<a/> | ||
==References== | ==References== | ||
1. Y. Chen, J. M. L. Ho, D. L. Shis, et al., Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 9, 1–8 (2018). | 1. Y. Chen, J. M. L. Ho, D. L. Shis, et al., Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 9, 1–8 (2018). | ||
Revision as of 13:17, 21 October 2021
Plux/lac_dE
Plux/lac_dE hybrid promotor
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 583
- 12INCOMPATIBLE WITH RFC[12]Illegal SpeI site found at 583
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 583
- 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 583
- 1000COMPATIBLE WITH RFC[1000]
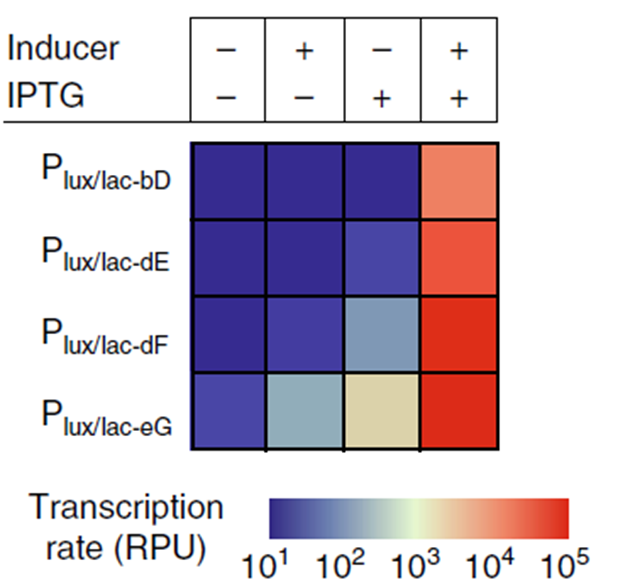
By combining multiple operator sites in a promoter region, a “hybrid promoter” can be created that responds to multiple input signals. This creates a logic transcriptional AND gate, of which the dynamic range can be further tuned by changing -35 and -10 sites. This E. coli promoter, Plux/lac, was developed by Chen et al. (2018) [1] and contains operator sites for LacI repression and LuxR activation. Transcription is fully activated when LacI is absent and LuxR is present. Different -10 sites “E” and “F” influence the transcription rate when only one of the two input signals is present (Figure 1).
<a href="https://2021.igem.org/wiki/index.php?title=Team:Wageningen"
>BB2<a/>
References
1. Y. Chen, J. M. L. Ho, D. L. Shis, et al., Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 9, 1–8 (2018).
