Difference between revisions of "Part:BBa K2165004"
| Line 143: | Line 143: | ||
<td><i>ura3-1::pYTK074-Integration-pCUP1-Venus-tENO1-URA3</i></td> | <td><i>ura3-1::pYTK074-Integration-pCUP1-Venus-tENO1-URA3</i></td> | ||
<td>Strain with Venus under CUP1 promoter, integration into <i>ura3-1</i> locus</td> | <td>Strain with Venus under CUP1 promoter, integration into <i>ura3-1</i> locus</td> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</table> | </table> | ||
| Line 165: | Line 152: | ||
===Microscopy=== | ===Microscopy=== | ||
Before microscopy, ET51, and ET52 strains were grown in 3 ml of uracil-dropout CSM (Complete Supplement Mixture, 100 mM MES buffer (2-ethanesulfonic acid diluted in 6M NaOH till pH=5.5); 2% glucose) for 3 hours at 30˚C. After that, every strain culture was split into two: in one 300 µM CuSO<sub>4</sub> was added and the second culture was used as a no-induction control. Cultures were grown for another 3 hours to OD<sub>600</sub> 0.2-0.8. | Before microscopy, ET51, and ET52 strains were grown in 3 ml of uracil-dropout CSM (Complete Supplement Mixture, 100 mM MES buffer (2-ethanesulfonic acid diluted in 6M NaOH till pH=5.5); 2% glucose) for 3 hours at 30˚C. After that, every strain culture was split into two: in one 300 µM CuSO<sub>4</sub> was added and the second culture was used as a no-induction control. Cultures were grown for another 3 hours to OD<sub>600</sub> 0.2-0.8. | ||
| − | |||
After that, 0.5 µl of cell culture was pipetted onto a 0.08 mm cover glass slip and covered with 1.5% agar-CSM (low melting temperature agarose was used). Zeiss Observer Z1 microscope with an automated stage, 63C/1.4NA oil immersion objective, and Axiocam 506 mono camera was used for imaging. During imaging, the focus was kept using Definite Focus and the sample was kept at 30°C using PeCon TempControl 37-2. ImageJ was used for image processing. | After that, 0.5 µl of cell culture was pipetted onto a 0.08 mm cover glass slip and covered with 1.5% agar-CSM (low melting temperature agarose was used). Zeiss Observer Z1 microscope with an automated stage, 63C/1.4NA oil immersion objective, and Axiocam 506 mono camera was used for imaging. During imaging, the focus was kept using Definite Focus and the sample was kept at 30°C using PeCon TempControl 37-2. ImageJ was used for image processing. | ||
| Line 178: | Line 164: | ||
<img style="width:100%;" src="https://2021.igem.org/wiki/images/0/0a/T--Estonia_TUIT--Contribution_CUP1.jpg"><br><br> | <img style="width:100%;" src="https://2021.igem.org/wiki/images/0/0a/T--Estonia_TUIT--Contribution_CUP1.jpg"><br><br> | ||
<i><b>Figure 1. Characterisation of pCUP1 | <i><b>Figure 1. Characterisation of pCUP1 | ||
| − | </b></I>Expression from CUP1 promoter is tightly controlled. The activity of the promoter was measured by quantifying the fluorescence intensity of Venus that is expressed from the | + | </b></I>Expression from CUP1 promoter is tightly controlled. The activity of the promoter was measured by quantifying the fluorescence intensity of Venus that is expressed from the promoter. The bars show mean fluorescence signals, error bars show standard deviation. |
</figure> | </figure> | ||
</center> | </center> | ||
Latest revision as of 09:22, 21 October 2021
CUP1 yeast inducible promoter with RFC[10] restriction sites removed
The Cup1 promoter enables binding of RNA Polymerase II and the subsequent transcription of downstream DNA to mRNA. It is activated by ACE1, a transcription factor which binds to copper ions. It was previously available as a standalone part as BBa_K945002, produced by Tec-Monterrery’s 2012 iGEM team; however, the original part contained numerous illegal restriction sites preventing its usability in standard biobrick assembly. Hard information of the Greenfield 2011 team’s BBa_K586000 claims the part is the Cup1 regulatory region, but based on the sequence and its twin we believe it to be mislabeled and that it is instead the CupI CDS.
We acquired our starting sequence from the Vicker’s Lab at the Australian Institute for Bioengineering and Nanotechnology via Addgene, subsequently removed or altered bases corresponding to illegal cut sites, added a biobrick prefix and suffix, and finally ordered the unit as a geneblock from IDT.
The followings were edited by iGEM17_Tianjin
Information Supplement
Transcription with CUP1 promoter is activated by sequence-specific transcription factors which bind to the CUP1 upstream activating sequence (CUP1 UAS). This sequence contains five binding sites for the Cu2+-dependent transcriptional activator ACE1/CUP2. The CUP1 UAS is necessary and sufficient for rapid and robust activation of this transcription.
The activation process is related to the acetylation of H3 and H4 located at CUP1 promoter, which showed nucleosome reposition and transcription factors binding might be the main reason for the activation.
Characterization
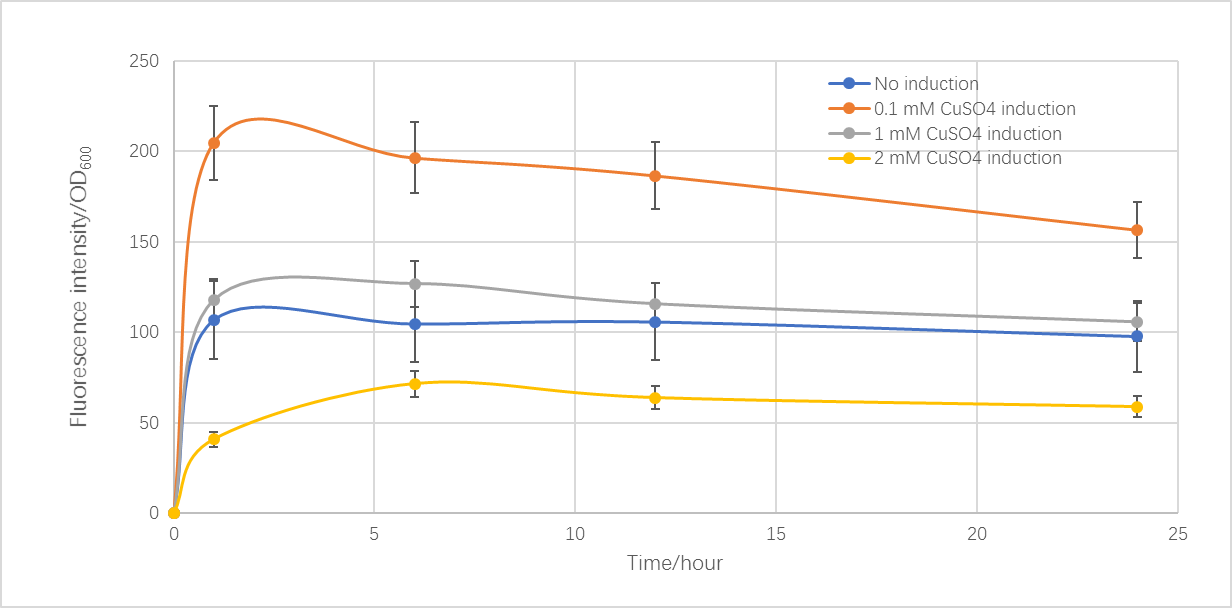
Based on the CUP1 promoter (BBa_K2165004) provided by iGEM16_Washington, we constructed a biosensor with yEmRFP. To characterize this promoter, strains of S. cerevisiae BY4742 containing the plasmid with an initial OD600 of 0.1 were grown for 24 hours in SC-URA medium at 30 degrees Celsius, and then were induced with copper sulfate. Samples in different copper concentration were tested with fluorescent microplate reader after 1, 6, 12, and 24 hours. This protocol was based on the experience used by Waterloo and Washington iGEM teams and amended by our team.
Figure 1 showed the relationship between fluorescence intensity with induction time and Cu2+ concentration. With 0.1 mM CuSO4 induced, the fluorescence intensity is 2 times over a control with no induction at 1 hour. As time went on, the fluorescence intensity slightly reduced. Moreover, as the Cu2+ concentration increased, the fluorescence intensity decreased, and when the concentration reached 1 mM, the intensity was close to the control group. This might be due to the higher copper ion concentration influences the transcription, expression and even growth of yeast.
Improvement
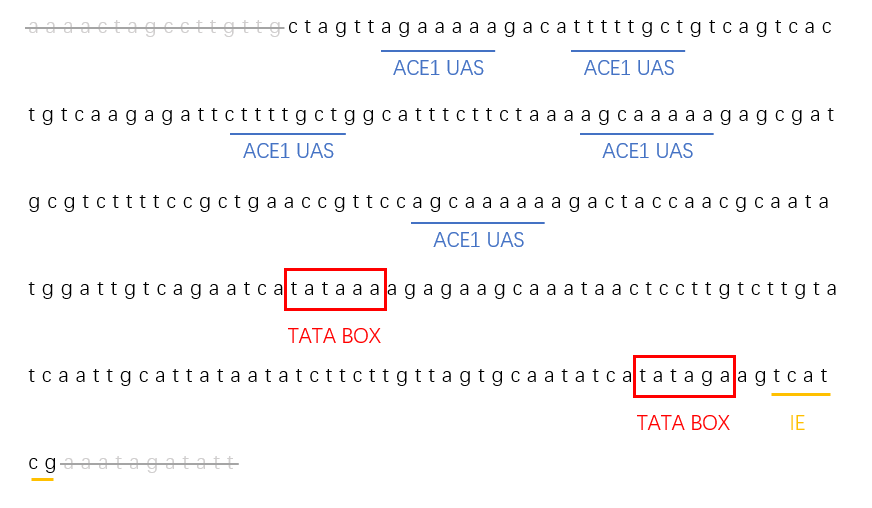
Based on this BioBrick, we redesigned the part sequence provided by iGEM16_Washington. We deleted irrelevant bases on the two ends of this promoter and retained the core sequence. We retained 5 ACE1/CUP1 binding sites, 2 TATA boxes, and one initiation element in the promoter. The complex of ACE1 and copper ions will bind the promoter, which causes the activation of CUP1 promoter with TATA boxes' help. In this way, this promoter played its key role with less bases.
Visit BBa_K2407000 to see more information.
Actually, we built some new parts based on CUP1 promoter by error-prone PCR. Visit BBa_K2407013, BBa_K2407014, BBa_K2407015, and BBa_K2407016 to get more information about characterization.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Estonia_TUIT 2021 team contribution
CUP1 promoter
CUP1 promoter regulates the expression of the CUP1 gene encoding for metallothioneins, molecules responsible for metal binding and controlling metal toxicity (Wang et al., 2016b). S. cerevisiae CUP1 promoter is a copper-inducible promoter. Copper ions bind to the N-terminal domain of the transcriptional activator Ace1 to cause a conformational change that triggers recognition of the activating sequences in the CUP1 promoter. Transcriptional activation occurs through the C-terminus of Ace1. CUP1 can also be induced with heat shock instead of copper ions (copper-independent pathway, (Leblanc et al., 2000b)).
Plasmid construction
To characterise the CUP1 promoter, we constructed plasmids using Golden Gate assembly and the MoClo yeast toolkit parts (Lee et al., 2015). Two plasmids were designed: low-copy number (CEN6/ARS origin of replication) and integrative plasmids (URA 3'-homology). We cloned the gene encoding for the Venus fluorescent reporter protein to these vectors under the control of our target promoter. The idea behind that is to detect, quantify and compare Venus fluorescence signals from plasmids with different copy numbers. The main features of the constructed plasmids are shown in Table 1.
Table 1. The main features of constructed plasmids.
| Promoter | Reporter | Origin of replication | Yeast integration site | Yeast selection gene | Copy number |
| pCUP1 | Venus | CEN6/ARS4 | - | URA3 | Low |
| pCUP1 | Venus | - | URA 3'-homology | URA3 | Low |
Yeast strain construction
Constructed integration vectors were restricted with NotI and used for transformation of S. cerevisiae DOM90 (MATα {leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 bar1::hisG} [phi+]) strain. The CEN6/ARS4 plasmids were used without linearization. Transformants were selected for URA+ phenotype on uracil-dropout CSM plates containing 2% glucose. All yeast strains generated and used for promoter characterisation are listed in Table 2.
Table 2. Yeast strains used for promoter characterisation.
| Strain name | Genotype | Description |
| DOM90 | MATα {leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 bar1::hisG} [phi+] | Background strain without Venus (control) |
| ET51 | pYTK074-CEN-pCUP1-Venus-tENO1-URA3 | Strain with Venus under CUP1 promoter, low copy number |
| ET52 | ura3-1::pYTK074-Integration-pCUP1-Venus-tENO1-URA3 | Strain with Venus under CUP1 promoter, integration into ura3-1 locus |
Microscopy
Before microscopy, ET51, and ET52 strains were grown in 3 ml of uracil-dropout CSM (Complete Supplement Mixture, 100 mM MES buffer (2-ethanesulfonic acid diluted in 6M NaOH till pH=5.5); 2% glucose) for 3 hours at 30˚C. After that, every strain culture was split into two: in one 300 µM CuSO4 was added and the second culture was used as a no-induction control. Cultures were grown for another 3 hours to OD600 0.2-0.8.
After that, 0.5 µl of cell culture was pipetted onto a 0.08 mm cover glass slip and covered with 1.5% agar-CSM (low melting temperature agarose was used). Zeiss Observer Z1 microscope with an automated stage, 63C/1.4NA oil immersion objective, and Axiocam 506 mono camera was used for imaging. During imaging, the focus was kept using Definite Focus and the sample was kept at 30°C using PeCon TempControl 37-2. ImageJ was used for image processing.
Results
We characterised the expression from the CUP1 promoter using Venus fluorescent protein as the reporter gene. The expression cassettes were either integrated to the yeast genome or on a centromeric low-copy plasmid, and the Venus expression was monitored in the presence and absence of the induction. CUP1 promoter was induced by addition of CuSO4. The experiments confirmed the studied promoter is strongly regulated by the presence of the induction, as the Venus fluorescence intensity increased drastically upon induction (Figure 1). The expression from the chromosomal locus was lower than from plasmid. These experiments confirm that CUP1 is a tightly-regulated promoter that mediates strong expression of the gene in the presence of the inductor.
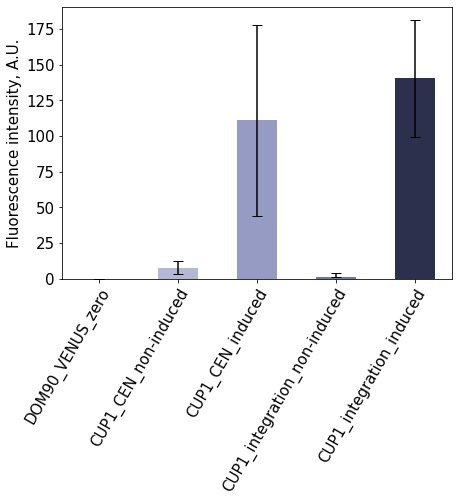
Figure 1. Characterisation of pCUP1 Expression from CUP1 promoter is tightly controlled. The activity of the promoter was measured by quantifying the fluorescence intensity of Venus that is expressed from the promoter. The bars show mean fluorescence signals, error bars show standard deviation.
References
- Wang, Y., Miao, X., Sun, J., & Cai, L. (2016b). Oxidative Stress in Diabetes: Molecular Basis for Diet Supplementation. Molecular Nutrition and Diabetes: A Volume in the Molecular Nutrition Series, 65–72. https://doi.org/10.1016/B978-0-12-801585-8.00006-3
- Leblanc, B. P., Benham, C. J., & Clark, D. J. (2000b). An initiation element in the yeast CUP1 promoter is recognized by RNA polymerase II in the absence of TATA box-binding protein if the DNA is negatively supercoiled. Proceedings of the National Academy of Sciences, 97(20), 10745–10750. https://doi.org/10.1073/PNAS.200365097
- Lee, M. E., DeLoache, W. C., Cervantes, B., & Dueber, J. E. (2015). A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synthetic Biology, 4(9), 975–986. https://doi.org/10.1021/SB500366V