Difference between revisions of "Part:BBa K4054009"
Shayne-Han (Talk | contribs) |
Shayne-Han (Talk | contribs) |
||
| Line 13: | Line 13: | ||
<p>Firstl, we use cell free and BL21 to express the RepA eGFP to confirm that the added tag will not affect the common function fo eGFP.</p> | <p>Firstl, we use cell free and BL21 to express the RepA eGFP to confirm that the added tag will not affect the common function fo eGFP.</p> | ||
[[File:T--XJTLU-CHINA--eGFP.png|600px|thumb|center|alt=domestication.|Figrure 2.The result of real-time detection while the eGFP expression was reduced by different concentration of LuxR.]] | [[File:T--XJTLU-CHINA--eGFP.png|600px|thumb|center|alt=domestication.|Figrure 2.The result of real-time detection while the eGFP expression was reduced by different concentration of LuxR.]] | ||
| − | <P>As Figure 1 shows, the expression product performs a peak of emission around 488 nm and a peak of excitation around 510 nm, which | + | <P>As Figure 1 shows, the expression product performs a peak of emission around 488 nm and a peak of excitation around 510 nm, which indicated that the RepA-eGFP was sucucessfully expressed while it remain the original charasteristics of none-tagged eGFP.</P> |
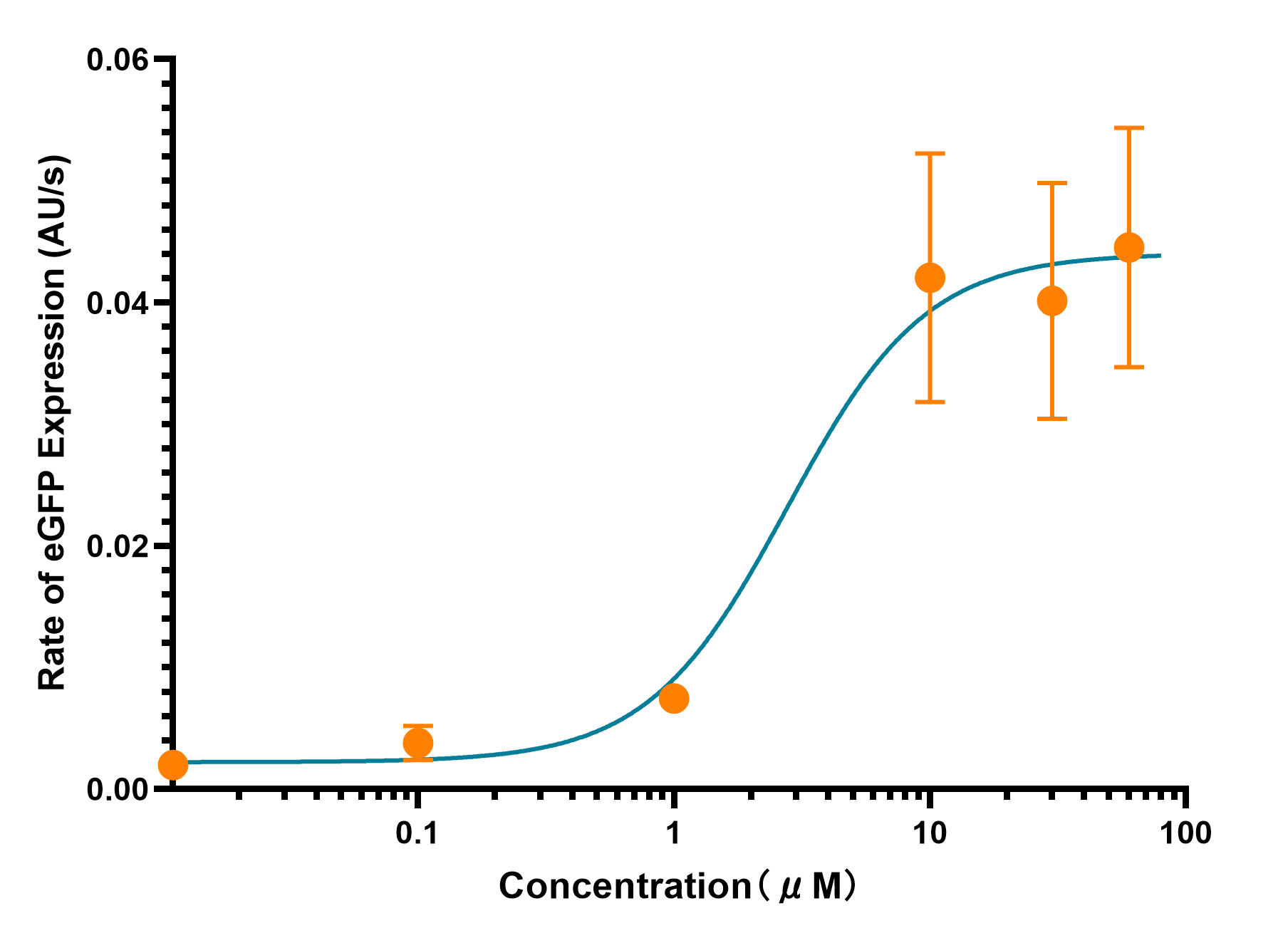
[[File:T--XJTLU-CHINA-S Curve.png|600px|thumb|center|alt=domestication.|Figure 3. The trend of expression rate with the changes of LuxR concentration.]] | [[File:T--XJTLU-CHINA-S Curve.png|600px|thumb|center|alt=domestication.|Figure 3. The trend of expression rate with the changes of LuxR concentration.]] | ||
<!-- --> | <!-- --> | ||
Revision as of 11:07, 20 October 2021
eGFP(enhanced green fluorescent protein) with N-terminal degradation tag RepA
In this year, we designed a composite part for adding the self-degration function to eGFP by bind a 16-amino-acid long tag called replication protein A(RepA) at the N-terminal of eGFP. The expressed eGFP will be continuely degraded by ClpAP in vivo or in vitro. Therefore, the moifed eGFP will be more specifcally reflect the state of promoter, when it works ad a repoter gene.
For us, we designed this part, RepA-eGFP, for observe the signal in real time and improve the specificity of our detecton, as they will not accumulate in our system. It also provide the availible method to improve the LuxR (an activator) we used in our project, to form a self-regulated device.
Usage and Biology
eGFP (enhanced green flourescent protein) is commonly used as a reporter in several quilified ot quantified meansurement. RepA tag could be used for increase the speed of degradation of teagged protein. It helps the recognation by ClpA enzyme and then ClpA could send the specific protein to ClpP for degradation. Meanwhile. the N-terminal tagged could avoid the structual effect around the C-terminal.
For us, we design a xomposite part for RepA-eGFP experssion in vivo, we chose the BL21(DE3) as the chassis and which contains the gene coding ClpAP, which means it could rapidly degrade the RepA-eGFO in cells.
Firstl, we use cell free and BL21 to express the RepA eGFP to confirm that the added tag will not affect the common function fo eGFP.
As Figure 1 shows, the expression product performs a peak of emission around 488 nm and a peak of excitation around 510 nm, which indicated that the RepA-eGFP was sucucessfully expressed while it remain the original charasteristics of none-tagged eGFP.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]