Difference between revisions of "Part:BBa K4000001"
| Line 16: | Line 16: | ||
=== Experimental approach === | === Experimental approach === | ||
1.GA plasmids transformation | 1.GA plasmids transformation | ||
| − | [[File:T--Fujian United--BBa K4000001 Figure20.jpg|500px|thumb|center| | + | [[File:T--Fujian United--BBa K4000001 Figure20.jpg|500px|thumb|center|Figure 2. GA plasmids transformed Saccharomyces cerevisiae..]] |
GA-expressing plasmids transformation and positive S. cerevisiae transformants selection using 50 μg/mL (Figure 2A) and 350 μg/mL (Figure 2B) hygromycin. Top left, pYES2-ctl. Top right, pYES2-TGC. Bottom left, pYES2-HGC. Bottom right, pYES2-IGC. | GA-expressing plasmids transformation and positive S. cerevisiae transformants selection using 50 μg/mL (Figure 2A) and 350 μg/mL (Figure 2B) hygromycin. Top left, pYES2-ctl. Top right, pYES2-TGC. Bottom left, pYES2-HGC. Bottom right, pYES2-IGC. | ||
| Line 22: | Line 22: | ||
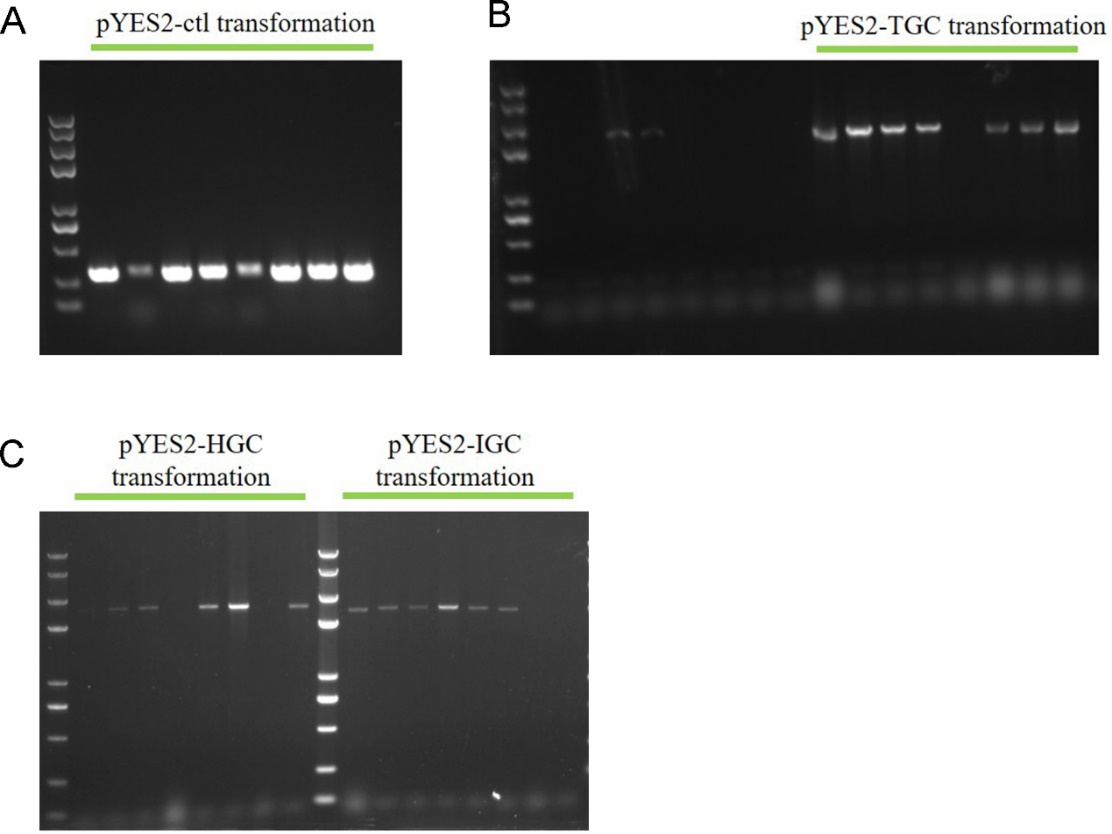
2. Colony PCR to identify the correct transpormed S. cerevisiae | 2. Colony PCR to identify the correct transpormed S. cerevisiae | ||
| − | [[File:T--Fujian United--BBa K4000001 Figure3.png|500px|thumb | + | [[File:T--Fujian United--BBa K4000001 Figure3.png|500px|thumb|Figure 3. Verification of the plasmids pYES2-ctl (A), pYES2-TGC (B), pYES2-HGC and pYES2-IGC (C) transformation via colony PCR in CEN.PK2-1C strain..]] |
Revision as of 07:14, 20 October 2021
GA
Profile
Name: GA
Base Pairs: 1567bp
Origin: Saccharomycopsis fibuligera, synthesis
Properties: An enzyme that can easily break down starches into glucose
Usage and Biology
Glucoamylase is an enzyme that can be obtained from the yeast or fungi in the Aspergillus genus such as Aspergillus niger. The enzyme decomposes starch molecules in the human body into the useful energy compound of glucose. This is accomplished by removing the alpha-1 and 4-glycosidic linkages from the non-reducing end of the starch molecule. These molecules are more commonly referred to as polysaccharides and are frequently either amylase- or amylopectin-based.
Experimental approach
1.GA plasmids transformation
GA-expressing plasmids transformation and positive S. cerevisiae transformants selection using 50 μg/mL (Figure 2A) and 350 μg/mL (Figure 2B) hygromycin. Top left, pYES2-ctl. Top right, pYES2-TGC. Bottom left, pYES2-HGC. Bottom right, pYES2-IGC.
2. Colony PCR to identify the correct transpormed S. cerevisiae
The colonies which grew on the high concentration hygromycin plates were subjected to colony PCR to verify the plasmids transformation again. From Fig. 3 we can see that positive bands implied the plasmids transformation successfully.
Proof of function
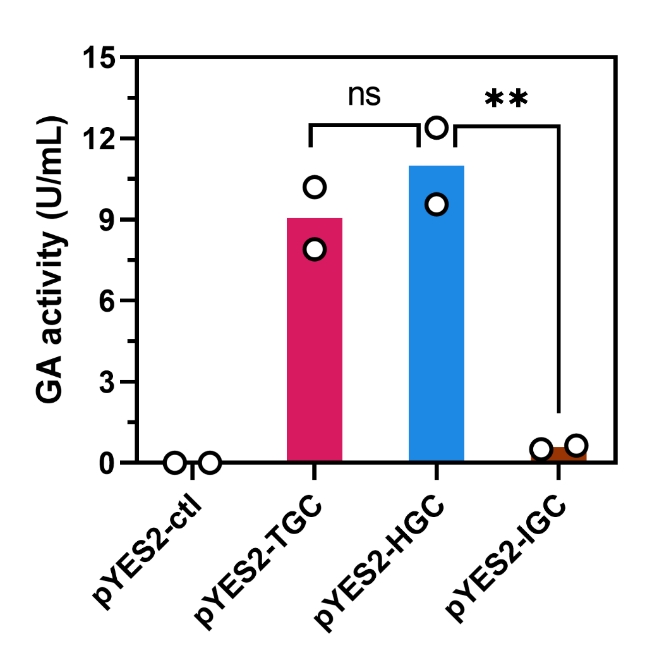
1. Glucoamylase activity assay
S. cerevisiae strains harboring various plasmids glucoamylase activity determination. **Statistical significance between indicated strains by Student’s t test, p < 0.01. ns, not significant. Data represent the means of two independent colonies.
2. Fermentation test
To verify the GA secretion capacity of the GA-expressing S. cerevisiae strains, we measured the glucose concentration inside the cell during the corn starch fermentation process. At the initial stage (0 h), when the GA was added during the “starch-to-glucose” process, higher contents of glucose were detected than the process without GA addition.
References
1. Görgens J F, Bressler D C, van Rensburg E. Engineering Saccharomyces cerevisiae for direct conversion of raw, uncooked or granular starch to ethanol[J]. Critical reviews in biotechnology, 2015, 35(3): 369-391.
2. Van Zyl W H, Bloom M, Viktor M J. Engineering yeasts for raw starch conversion[J]. Applied microbiology and biotechnology, 2012, 95(6): 1377-1388.
3. Maury J, Kannan S, Jensen N B, et al. Glucose-dependent promoters for dynamic regulation of metabolic pathways[J]. Frontiers in bioengineering and biotechnology, 2018, 6: 63.
4. Weber E, Engler C, Gruetzner R, et al. A modular cloning system for standardized assembly of multigene constructs[J]. PloS one, 2011, 6(2): e16765.
5. Pollak B, Cerda A, Delmans M, et al. Loop assembly: a simple and open system for recursive fabrication of DNA circuits[J]. New Phytologist, 2019, 222(1): 628-640.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 131
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 92
Illegal AgeI site found at 1463 - 1000COMPATIBLE WITH RFC[1000]