Difference between revisions of "Part:BBa K4096004"
| Line 1: | Line 1: | ||
| + | == Profile == | ||
| + | === Name: pET-25b-PKC-OP === | ||
| + | === Base Pairs: 6633 bp === | ||
| + | === Origin: Synthetic === | ||
| + | === Properties: A recombinant plasmid containing cellulase sequence. === | ||
| − | + | == Usage and Biology == | |
| − | + | BBa_K4096004 is a plasmid that can expressed cellulase (PKC-OP) under the control of T7 promoter from Pseudomonas aeruginosa. Cellulases break down the cellulose molecule into monosaccharides ("simple sugars") such as beta-glucose, or shorter polysaccharides and oligosaccharides. Several different kinds of cellulases are known, which differ structurally and mechanistically. Synonyms, derivatives, and specific enzymes associated with the name "cellulase" include endo-1,4-beta-D-glucanase (beta-1,4-glucanase, beta-1,4-endoglucan hydrolase, endoglucanase D, 1,4-(1,3,1,4)-beta-D-glucan 4-glucanohydrolase), carboxymethyl cellulase (CMCase), avicelase, celludextrinase, cellulase A, cellulosin AP, alkali cellulase, cellulase A 3, 9.5 cellulase, and pancellase SS. | |
| − | pET-25b- | + | == Construct design == |
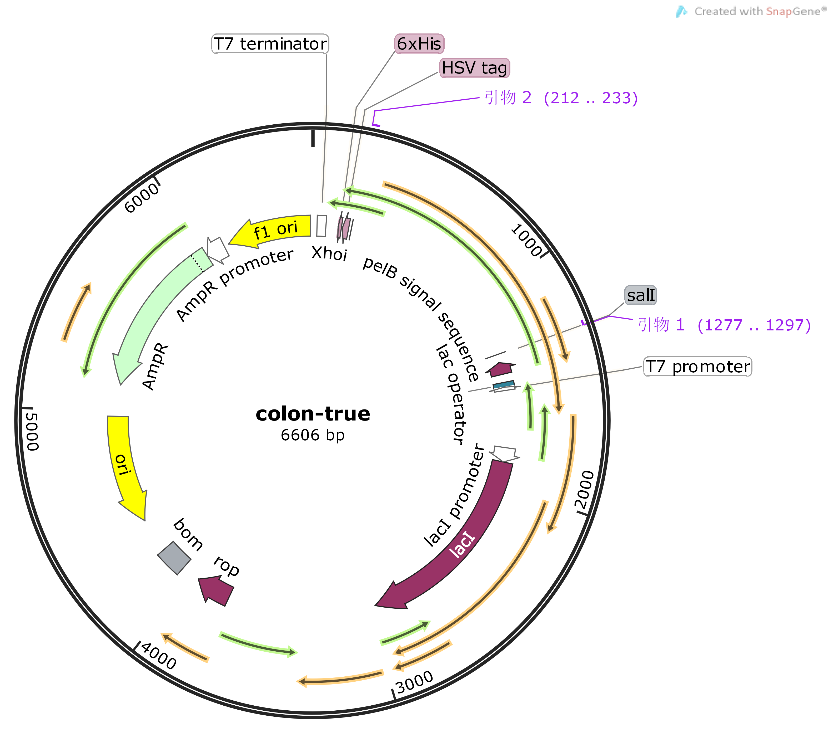
| + | The alkaline cellulase gene from Pseudomonas aeruginosa PKC-OP performed codon optimization on PKC-001 was selected and inserted into the pET-25b vector which contains a pelB signal peptide (Figure 1 and 2). The recombinant plasmid then was transformed it to E. coli BL21(DE3) strain to produce cellulase. | ||
| + | [[File:T--Shanghai Metro HS--BBa K4096002-Figure1.png|500px|thumb|center|Figure 1. Genetic design of our gene.]] | ||
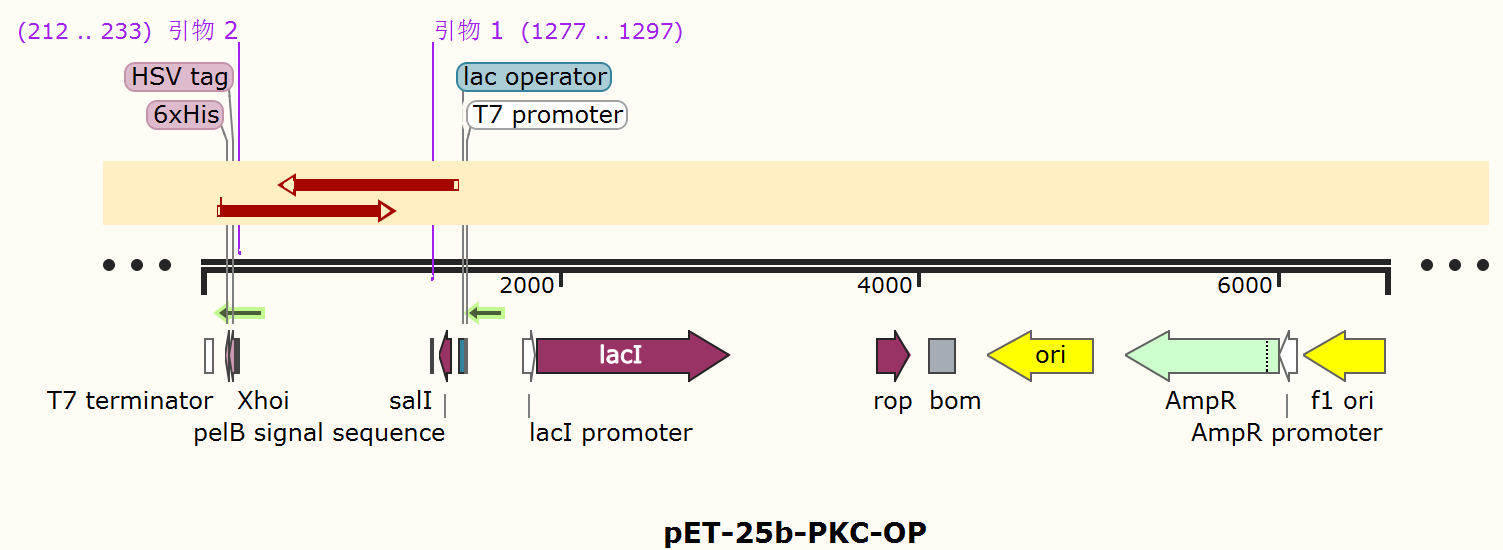
| + | [[File:T--Shanghai Metro HS--BBa K4096004-Figure2.png|500px|thumb|center|Figure 2. Schematic map of pET-25b-PKC expression plasmid..]] | ||
| + | The profiles of every basic part are as follows: | ||
| − | + | === BBa_K4096002 === | |
| − | === | + | ==== Name: PKC-OP ==== |
| + | ==== Base Pairs: 1086bp ==== | ||
| + | ==== Origin: Pseudomonas aeruginosa, genome ==== | ||
| + | ==== Properties: A coding sequence of alkali cellulase. ==== | ||
| − | + | === Usage and Biology === | |
| − | + | BBa_K4096002 is a coding sequence of alkali cellulase (PKC-OP) from Pseudomonas aeruginosa. Cellulases break down the cellulose molecule into monosaccharides ("simple sugars") such as beta-glucose, or shorter polysaccharides and oligosaccharides. Several different kinds of cellulases are known, which differ structurally and mechanistically. | |
| − | + | ||
| + | === BBa_K4096000 === | ||
| + | ==== Name: pET-25b-vector ==== | ||
| + | ==== Base Pairs: 5547bp ==== | ||
| + | ==== Origin: Addgene ==== | ||
| + | ==== Properties: A plasmid expressing proteins. ==== | ||
| − | + | ==== Usage and Biology ==== | |
| − | === | + | BBa_K4096000 is a plasmid that can express proteins. |
| − | + | ||
| − | + | == Experimental approach == | |
| + | Construction of recombinant plasmid | ||
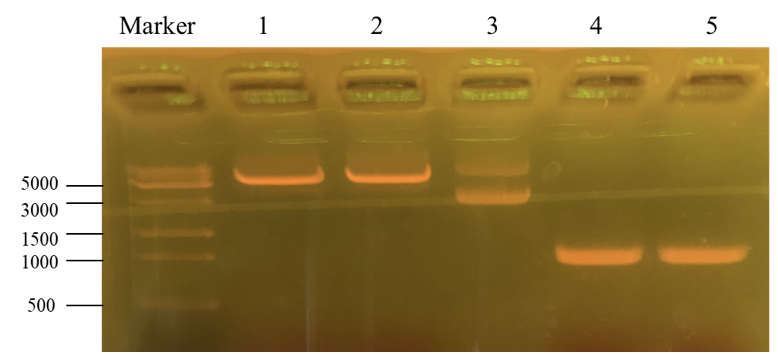
| + | [[File:T--Shanghai Metro HS--BBa K4096002-Figure2.png|500px|thumb|center|Figure 3. The result of enzyme cutting and PCR.]] | ||
| + | This picture is the nucleic acid electrophoresis result of enzyme cutting and PCR (By performing PCR, we can obtain the desirable PKC-OP’s gene segment.). Column “marker” is a column that is used to show the position of different lengths of genes. In this step, our aim is to verify whether these results are the desired ones. | ||
| + | Number 1 and 2 is the result for pET-25b after enzyme digestion. Additionally, number 3 is the pET-25b without enzyme cutting. At last, number 4 and 5 is the PKC-OP after PCR. As our experiment moves on, the concentration of our PKC-OP and pET-25b are 233.2ng/μL and 17.8ng/μL. Lastly, we use the homologous combination to combine them. | ||
| + | |||
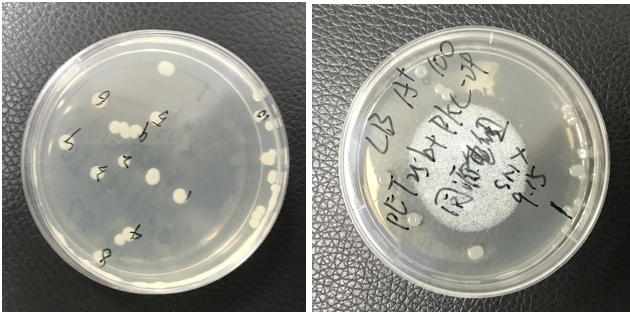
| + | [[File:T--Shanghai Metro HS--BBa K4096002-Figure3.png|500px|thumb|center|Figure 4 E. coil having the desired pET-25b and PKC-OP.]] | ||
| + | The pET25b-PKC-OP was constructed. | ||
| + | The plate shows monoclonals of pET25b-PKC-OP constructs. | ||
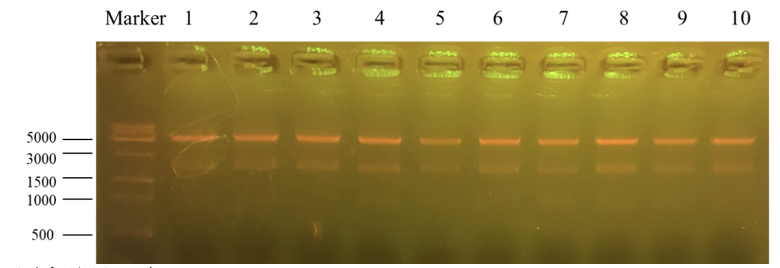
| + | [[File:T--Shanghai Metro HS--BBa K4096002-Figure4.png|500px|thumb|center|Figure 5. The result of Apa1 enzyme digestion validation.]] | ||
| + | Column “marker” is a column that is used to show the position of different lengths of genes. Number 1 to 10 is the result for recombinant plasmid pET25b-PKC-OP after Apa1 enzyme digestion. We get two bands of 4712bp and 1894bp. It further indicates that the obtained monoclonals were positive monoclonals containing the recombinant plasmid. | ||
| + | 1, 7 and 8 plasmids were sent to sequence. | ||
| + | |||
| + | |||
| + | [[File:T--Shanghai Metro HS--BBa K4096004-Figure6.png|500px|thumb|center|Figure 6 The result of sequencing for plasmid pET25b-PKC-OP..]] | ||
| + | Sequencing feedback shows we have obtained the correct plasmids which is consistent with their DNA profiles. | ||
| + | Different induction conditions were tested for protein expression. As the pET-25b vector contains a pelB signal peptide, the engineered strain would secret the protein into the medium. Therefore, we collected the culture supernatant after induction and ran SDS-PAGE for verification (Figure 2). | ||
| + | |||
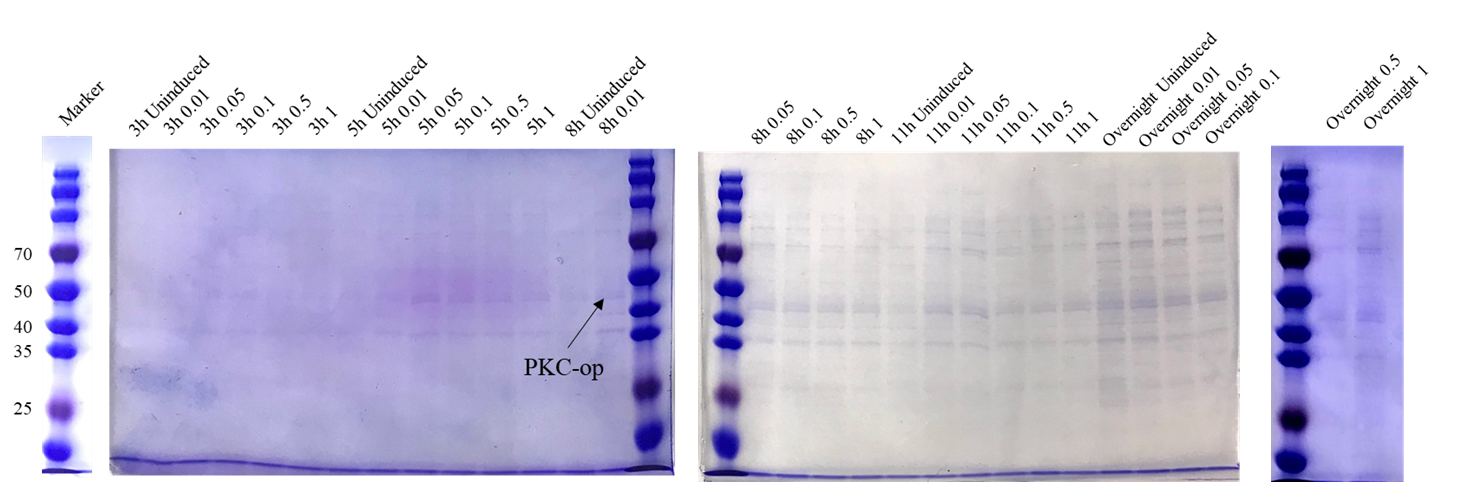
| + | [[File:T--Shanghai Metro HS--BBa K4096002-Figure5.png|500px|thumb|center|Figure 7. SDS-PAGE analysis of culture supernatant under different induction conditions..]] | ||
| + | The theoretical molecular weight of the PKC cellulase is 45.6 kDa. As seen from the SDS-PAGE (Figure 7), there is a wide band just above 40 kDa and it indicates that we have obtained the PKC cellulase. | ||
| + | |||
| + | == Proof of function == | ||
| + | As our goal is to produce the additive that could secret PKC enzyme to help degrade the cellulose in silage, we want to ensure that our engineered strain can make the best of itself at its best condition. From this perspective, we designed several different induction conditions for protein expression and decided to build the model to determine the optimal condition for our engineered strain to “work” better. | ||
Revision as of 08:42, 19 October 2021
Contents
Profile
Name: pET-25b-PKC-OP
Base Pairs: 6633 bp
Origin: Synthetic
Properties: A recombinant plasmid containing cellulase sequence.
Usage and Biology
BBa_K4096004 is a plasmid that can expressed cellulase (PKC-OP) under the control of T7 promoter from Pseudomonas aeruginosa. Cellulases break down the cellulose molecule into monosaccharides ("simple sugars") such as beta-glucose, or shorter polysaccharides and oligosaccharides. Several different kinds of cellulases are known, which differ structurally and mechanistically. Synonyms, derivatives, and specific enzymes associated with the name "cellulase" include endo-1,4-beta-D-glucanase (beta-1,4-glucanase, beta-1,4-endoglucan hydrolase, endoglucanase D, 1,4-(1,3,1,4)-beta-D-glucan 4-glucanohydrolase), carboxymethyl cellulase (CMCase), avicelase, celludextrinase, cellulase A, cellulosin AP, alkali cellulase, cellulase A 3, 9.5 cellulase, and pancellase SS.
Construct design
The alkaline cellulase gene from Pseudomonas aeruginosa PKC-OP performed codon optimization on PKC-001 was selected and inserted into the pET-25b vector which contains a pelB signal peptide (Figure 1 and 2). The recombinant plasmid then was transformed it to E. coli BL21(DE3) strain to produce cellulase.
The profiles of every basic part are as follows:
BBa_K4096002
Name: PKC-OP
Base Pairs: 1086bp
Origin: Pseudomonas aeruginosa, genome
Properties: A coding sequence of alkali cellulase.
Usage and Biology
BBa_K4096002 is a coding sequence of alkali cellulase (PKC-OP) from Pseudomonas aeruginosa. Cellulases break down the cellulose molecule into monosaccharides ("simple sugars") such as beta-glucose, or shorter polysaccharides and oligosaccharides. Several different kinds of cellulases are known, which differ structurally and mechanistically.
BBa_K4096000
Name: pET-25b-vector
Base Pairs: 5547bp
Origin: Addgene
Properties: A plasmid expressing proteins.
Usage and Biology
BBa_K4096000 is a plasmid that can express proteins.
Experimental approach
Construction of recombinant plasmid
This picture is the nucleic acid electrophoresis result of enzyme cutting and PCR (By performing PCR, we can obtain the desirable PKC-OP’s gene segment.). Column “marker” is a column that is used to show the position of different lengths of genes. In this step, our aim is to verify whether these results are the desired ones. Number 1 and 2 is the result for pET-25b after enzyme digestion. Additionally, number 3 is the pET-25b without enzyme cutting. At last, number 4 and 5 is the PKC-OP after PCR. As our experiment moves on, the concentration of our PKC-OP and pET-25b are 233.2ng/μL and 17.8ng/μL. Lastly, we use the homologous combination to combine them.
The pET25b-PKC-OP was constructed. The plate shows monoclonals of pET25b-PKC-OP constructs.
Column “marker” is a column that is used to show the position of different lengths of genes. Number 1 to 10 is the result for recombinant plasmid pET25b-PKC-OP after Apa1 enzyme digestion. We get two bands of 4712bp and 1894bp. It further indicates that the obtained monoclonals were positive monoclonals containing the recombinant plasmid. 1, 7 and 8 plasmids were sent to sequence.
Sequencing feedback shows we have obtained the correct plasmids which is consistent with their DNA profiles. Different induction conditions were tested for protein expression. As the pET-25b vector contains a pelB signal peptide, the engineered strain would secret the protein into the medium. Therefore, we collected the culture supernatant after induction and ran SDS-PAGE for verification (Figure 2).
The theoretical molecular weight of the PKC cellulase is 45.6 kDa. As seen from the SDS-PAGE (Figure 7), there is a wide band just above 40 kDa and it indicates that we have obtained the PKC cellulase.
Proof of function
As our goal is to produce the additive that could secret PKC enzyme to help degrade the cellulose in silage, we want to ensure that our engineered strain can make the best of itself at its best condition. From this perspective, we designed several different induction conditions for protein expression and decided to build the model to determine the optimal condition for our engineered strain to “work” better.