Difference between revisions of "Part:BBa K3264022"
(→SZU-China 2020 iGEM TEAM) |
|||
| Line 205: | Line 205: | ||
| + | ==<b>BJU-China 2021 iGEM TEAM</b>== | ||
| + | TnaA is endogenous tryptophanase from E. coli that can convert tryptophan to indole. MaFMO is monooxygenase from Methylophaga aminisulfidivorans which could effectively synthesize indigo by oxidation of indole. | ||
| + | ===Description=== | ||
| + | In our project, TnaA-MaFMO is a combination used in 6BrIG production strategy in E. coli BL21(DE3). Since E. coli inherently carries TnaA, to prevent the influence of endogenous expression, we knock out TnaA in the genome, and then compensate TanA on the pET28b plasmid together with MaFMO. The expression of TnaA-MaFMO was controlled by T7-LacO inducible promoter, and IPTG acts as inducer. | ||
| + | [[File:T--BJU_China--Fig.1 TanA-MaFMO1.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Figure 1. Gene circuit of TnaA-MaFMO</center></b></p> | ||
| + | |||
| + | ===Experiments and Results=== | ||
| + | We obtained TnaA gene and MaFMO gene by PCR and then assembled them to pET28b backbone by Gibson. Colony PCR and gene sequencing were use to verify the sequences. The following was the agarose gel electrophoresis of the results (Figure 2). | ||
| + | [[File:T--BJU_China--Fig.2 TanA-MaFMO2.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Figure 2. Agarose gel results of TnaA-MaFMO</center></b></p> | ||
| + | |||
| + | Next, we want to test the expression of these enzymes with IPTG induction. For better expression of enzymes, we optimize the induction condition of gene expression: three different temperatures (18℃, 30℃, 37℃) and inducer concentrations (0mM, 0.1mM, 1mM) were set for test. | ||
| + | We successfully detected TnaA-MaFMO enzyme after IPTG induction using SDS-PAGE (Figure 3). In the gel picture, we can clearly see TanA-MaFMO bands (about 52.8 KDa). And we can find that TanA-MaFMO expresses significantly better at 30℃ and 37℃ than 18℃. When the inducer concentration is lower (0.1mM), the protein bands are more obvious, and even when there is no inducer, the gene also have a basic expression, that is also called leakage. The leakage may caused by high gene expression using high copy plasmid pET28b. | ||
| + | [[File:T--BJU_China--Fig.3 TanA-MaFMO3.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Figure 3. SDS-PAGE results of TnaA-MaFMO</center></b></p> | ||
| + | |||
| + | In summary, we successfully constructed a plasmid including two enzymes TnaA and MaFMO, and detected their expression through the induction of IPTG. | ||
Revision as of 06:29, 19 October 2021
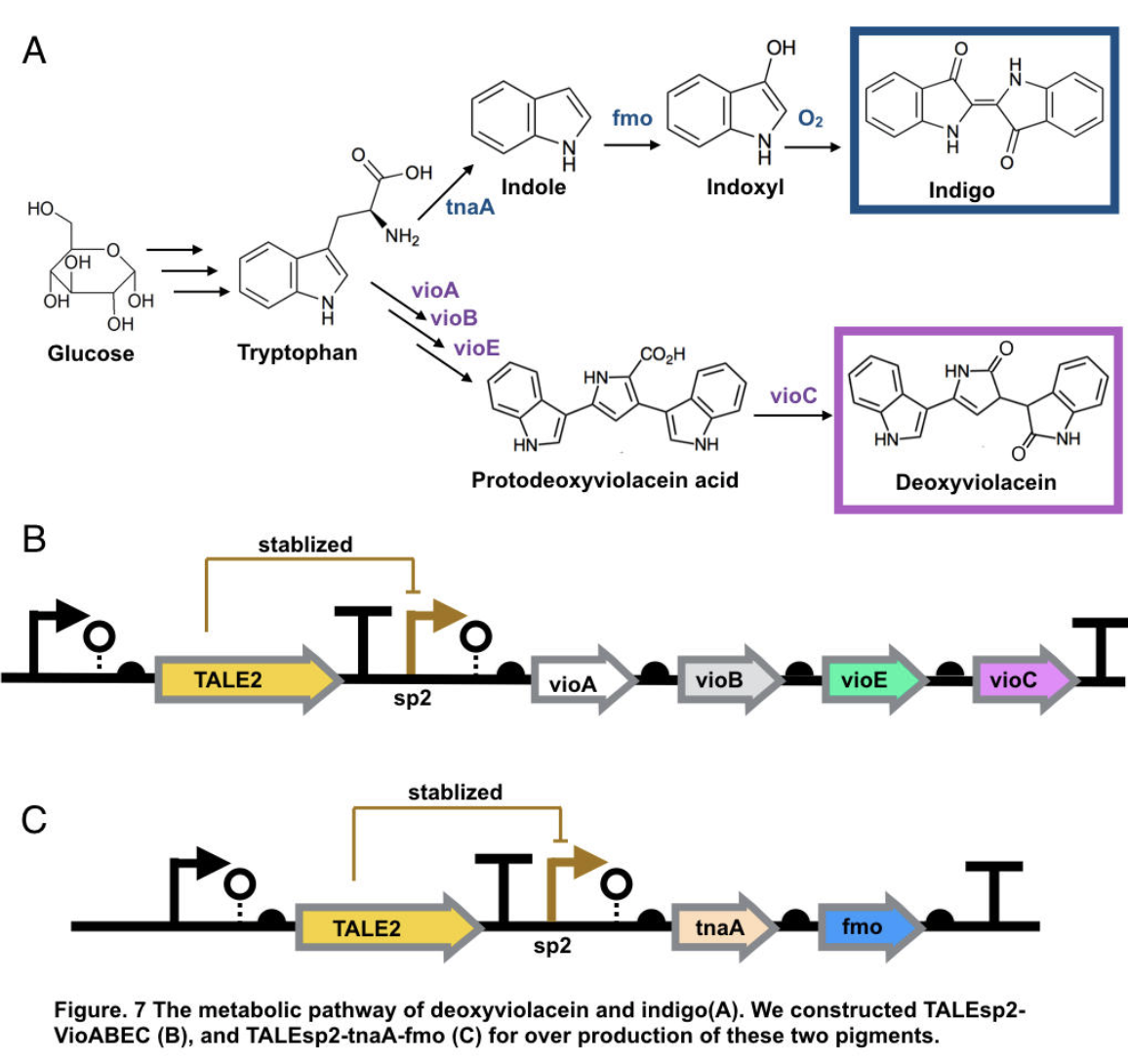
TALEsp2-tnaA-Fmo-UGT (rate by Talesp2)
Indigo is an ancient dye uniquely capable of producing the signature tones in blue denim; however, the dyeing process requires chemical steps that are environmentally damaging. We describe a sustainable dyeing strategy that not only circumvents the use of toxic reagents for indigo chemical synthesis but also removes the need for a reducing agent for dye solubilization. (FMO) from M. aminisulfidivorans can be expressed in many strains of Escherichia coli to produce indigo dye. In the presence of indole and oxygen, FMO can catalyze the addition of a hydroxyl group to indole generating the intermediate indoxyl. Indoxyl then naturally oxidizes to generate indigo.
Despite simply laying a complete database of spidroin, GreatBay_SZ this year also approached one major industry where spider silk held great potential: the cloth industry. Identifying the current chemical compound pollution during dying process as well as the damage brought by chemical fiber itself, we realized that the typically non-environmentally friendly material of cloth manufacture can be replaced by spider silk, as a thread to weaved the cloth, and natural dyes, as pigments that granted cloth color. Among these, we selected deoyviolacein (purple red color) and indigo (blue color), both natural dyes with admittedly strong stability and fascinating color tones [11], and shared a common precursor L-tyrptophan. Together, we hoped to weave a Spider-Man battle suits with our spider silk and dyes for demonstration.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1513
Illegal NheI site found at 1921 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 113
Illegal BamHI site found at 6028
Illegal XhoI site found at 2152
Illegal XhoI site found at 2692 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 747
Illegal AgeI site found at 849
Illegal AgeI site found at 1767
Illegal AgeI site found at 2277
Illegal AgeI site found at 4436 - 1000COMPATIBLE WITH RFC[1000]
Usage in biology
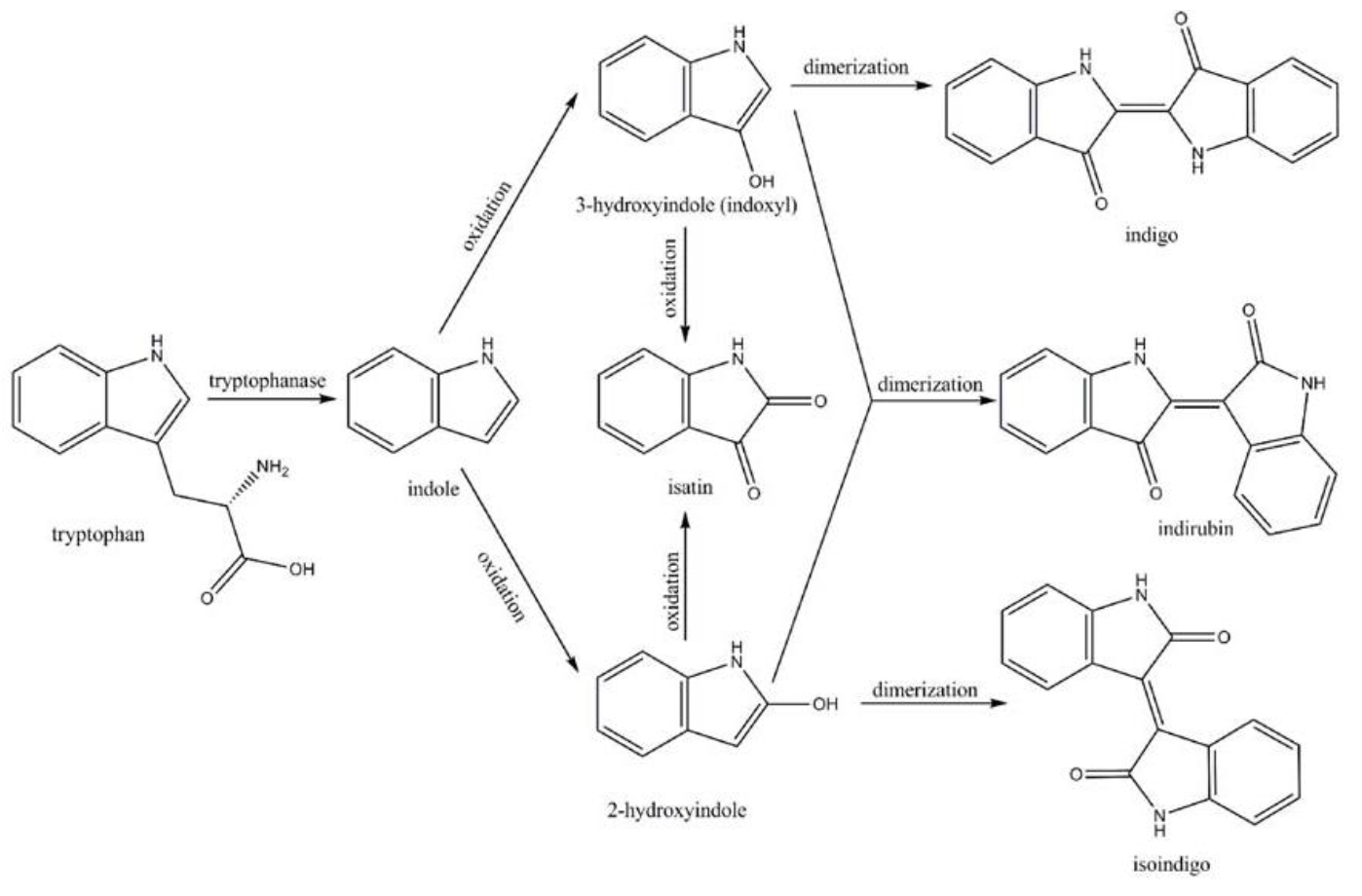
Indigo, though without much significant medical usage, is the most classic ancient dye uniquely capable of producing the signature tones in blue denim. The current production of indigo not only release hazardous chemical compound, but also requires an excess, usually toxic, reducing agent to reduce the insoluble indigo into soluble leucoindigo. Replacement of reducing agent were identified in the past, yet none established as fast nor cost effective. We therefore researched an alternative indigo production pathway. With L-tryptophan, indole is formed by enzyme tnaA in E.coli. In the presence of indole and oxygen, FMO catalyzes the addition of a hydroxyl group to indole generating the intermediate indoxyl that can spontaneously oxidize to form indigo.
The pigment deoxyviolacein is produced from L-tryptophan in E.coli via a pathway involving four enzymes VioA, VioB, VioE, VioC, only vioD is excluded through the pathway. GreatBay_SZ 2019 borrow from team SHSBNU_China to acquire part thsR- BBa_K274003, which include one thiosulfate sensor and vioABDE which synthesize proviolacein. As shown in the graph below, different arrangement of VioA-E can synthesize pigment with deviate color. Our team wants to substitute VioD to vioC that gives as a light pink/purple pigment
Aiming for stable and high efficient production through deoxyviolacein pathway, using the original thiosulfate sensor cannot yield us high concentration of pigment. Instead, we look into GreatBay_China_2018’s stabilized promoter BBa_K2753019, Talesp2 from the pTale family. Transcription-activator-like-effector (TALE) stabilised promoters are a type of promoters able to untie gene expression level from gene copy number using an incoherent feed forward loop (iFFL) in which transcription-activator-like effectors (TALEs) function as a perfectly non-cooperative negative regulation. While copy number accretes gene expression, it also elevates the repression to the gene expression, thus has canceled out the effect of copy number on expression level. Thus using Talesp2, we comprehend that it can yield high quantitative results
Characterization
To achieve over production of both pigments, we utilized the stabilized promoter BBa_K2753019 from GreatBay_China(2018), transcription-activator-like-effector (TALE) stabilised promoters that untie gene expression level from gene copy number. We designed to place TALEsp2 promoter on standard pSC101 backbones. Combining the pathway with promoter, we hence characterized and obtained pigments by constructing part TALEsp2-VioABEC for deoxyviolacein through BBa_K2753019, BBa_K726015, and a new basic part BBa_K3264008, a gene-transcript of VioC. Meanwhile, we also constructed a new composite part TALEsp2-tnaA-FMO for indigo.
Deoxyviolacein has long be identified as secondary metabolity actively against pathogenic bacteria like Pseudomonas aeruginosa and Staphylococcus aureus, and leukemia, lung cancer, human uveal melanoma, and lymphoma cells. It also served other purpose like natural pigments. The importance of violacein urged us to search for over production of both metabolities. The hidden pathway for production is encoded by the VioABCDE operon. Bio-synthesis starts from L-tryptophan, converted into protodeoxyviolaceinic acid by VioA, VioB and VioE enzymes, and then into deoxyviolacein is therefore produced with the activation of VioC gene[10]. All promoters are placed on the standard biobrick assembly compatible backbone pSC101. We reassembled plasmid thsR- BBa_K274003 by PCR vioAB, VioE and talesp2, only VioC was synthesized de novo by Genescript. All parts were assembled using Gibson Assembly.
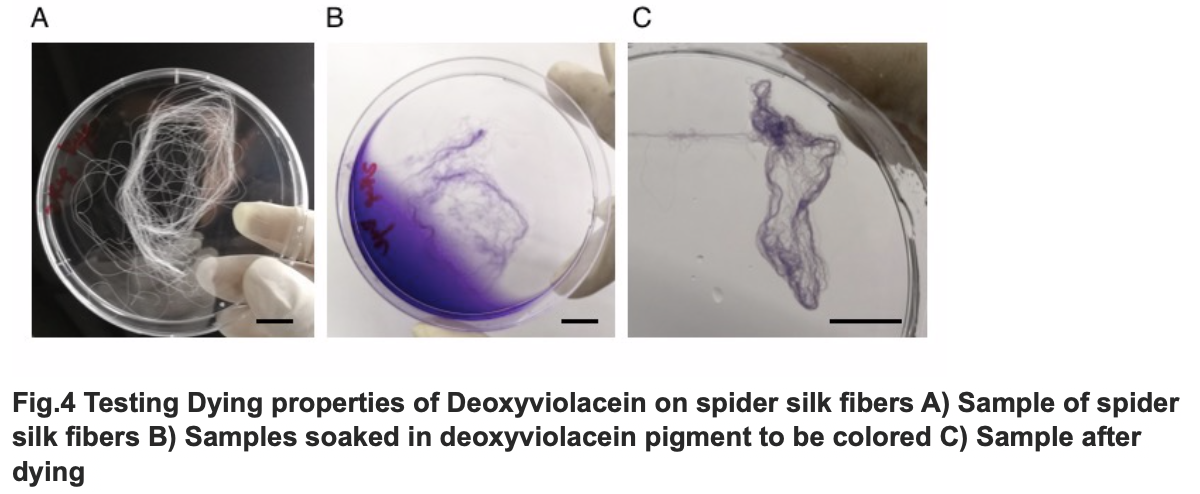
After we acquire pure extract deoxyviolacein pigment, we test it dying properties upon our spider silk sample. As documented as the process below, the pigment can be well soaked into fibers to give it light purple color.
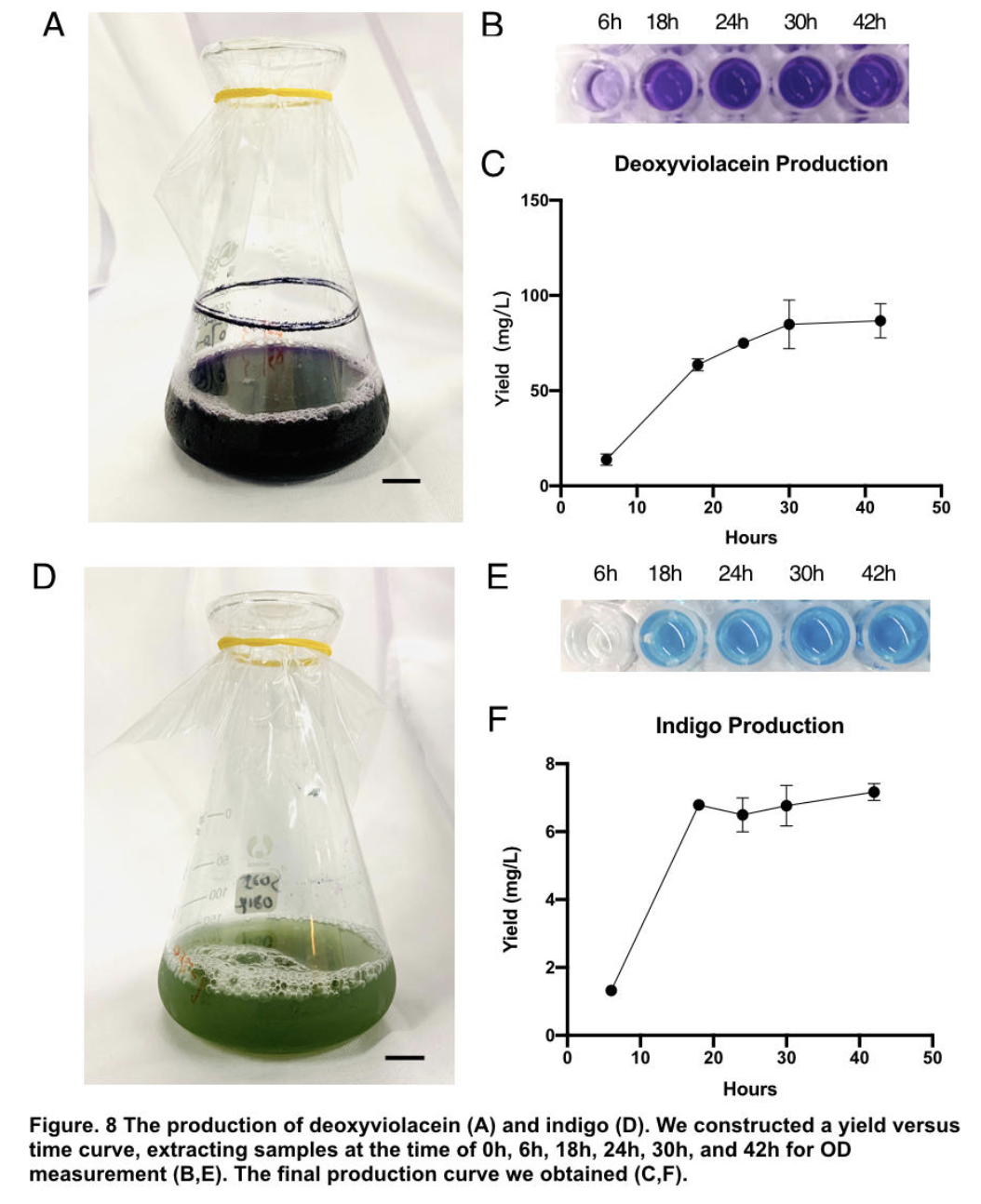
The influence of talesp2 on the production and accumulation of deoxyviolacein is remarkable, with the highest yield round 90~100mg/L which nearly matched the standard of pure extracts deoxyviolacein. It can be concluded that talesp2 has positive influences on deoxyviolacein’s metabolic reaction, which it stabilized towards an optimal level of gene expression that produce just enough enzyme to metabolize the substrate. After we acquire pure extract deoxyviolacein pigment, we test it dying properties upon our spider silk sample. As documented as the process below, the pigment can be well soaked into fibers to give it light purple color.
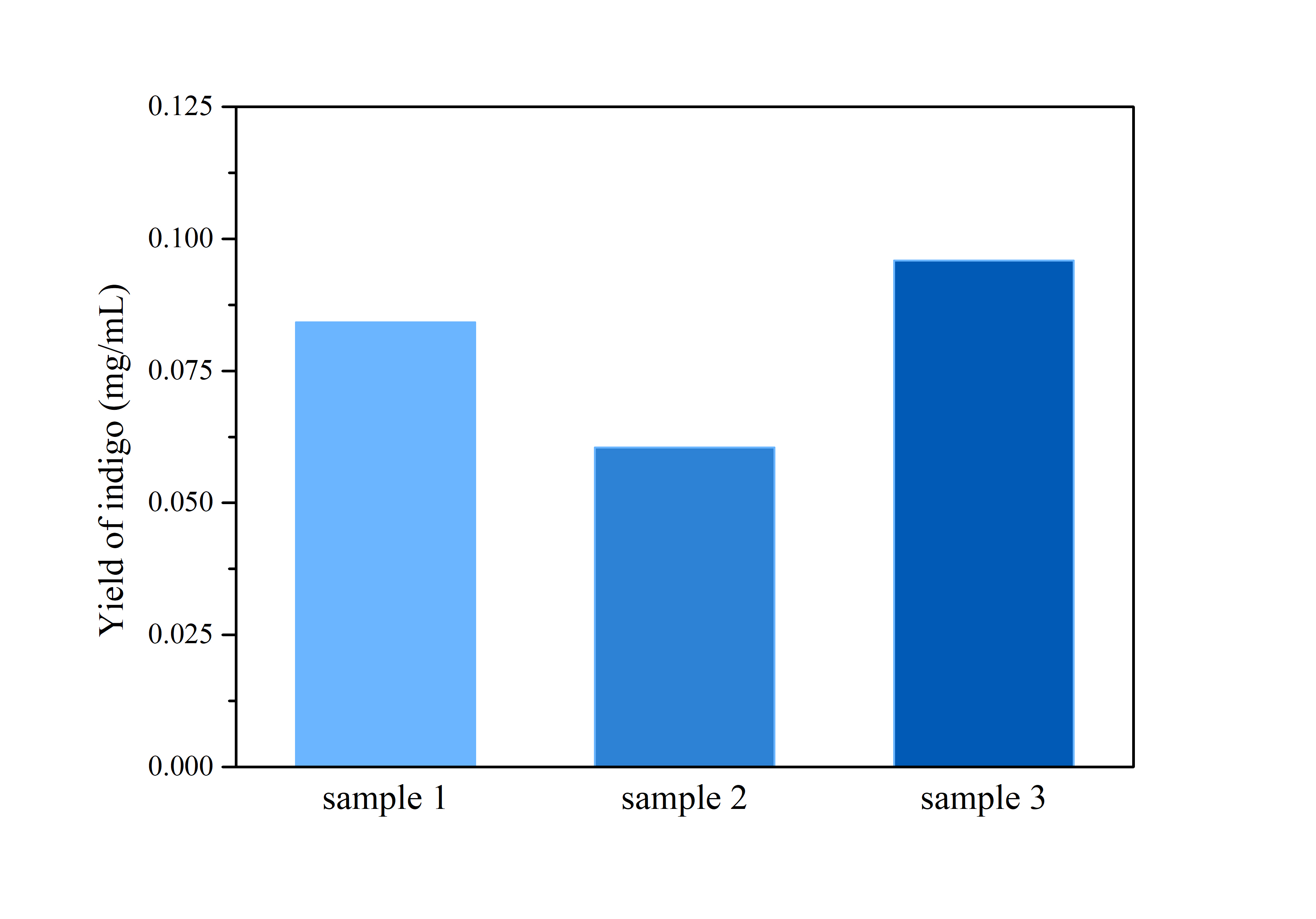
Pigments were obtained by extracting pigments after 42 hours of shake-flask incubation (without iptg) using solvent ethanol for violacein, and DMSO for indigo respectively. Through calculation based on standard indigo product and OD measurement of oh, 6h, 18h, 24h, 30h, 42h (peak of production), we are able to construct a yield versus time curve. Through that, we concluded our deoxyviolacein yields 85.81±9.09mg/L maximum and indigo yields 6.97±0.44mg/L maximum.
Testing Dying Properties on Spider silk Fibers
Reference
[1]Rodrigues, André L., et al. “Systems Metabolic Engineering of Escherichia Coli for Production of the Antitumor Drugs Violacein and Deoxyviolacein.”
Metabolic Engineering, vol. 20, 2013, pp. 29–41., doi:10.1016/j.ymben.2013.08.004.
[2]Wen, Rui, et al. “Molecular Cloning and Analysis of the Full-Length Aciniform Spidroin Gene from Araneus Ventricosus.” International Journal of Biological Macromolecules, vol. 117, 2018, pp. 1352–1360., doi:10.1016/j.ijbiomac.2017.12.090.
SZU-China 2020 iGEM TEAM
1.Species Source
In order to better characterize the bacterial flavin-containing monooxygenase (bFMO) documented by BBa_1131000, we found many literature about FMO that are also from M. aminisulfidivorans. The species’current name is Methylophaga aminisulfidivorans MP, and in some early literature it may be named as Methylophaga aminisulfidivorans SK1. Its taxonomy ID in NCBI is 1026882.
2.Experimental Characterization
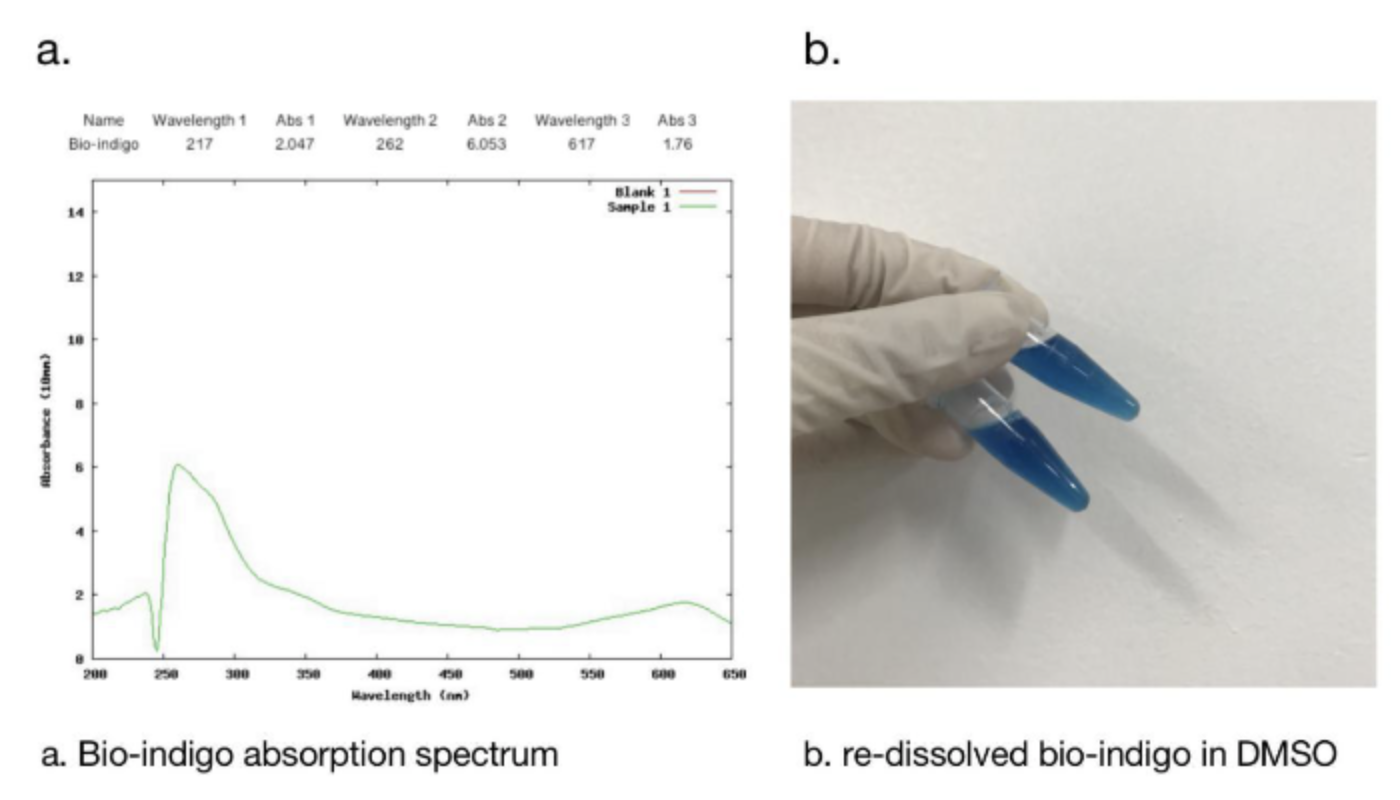
2.1 Bio-indingo Absorption Spectrum
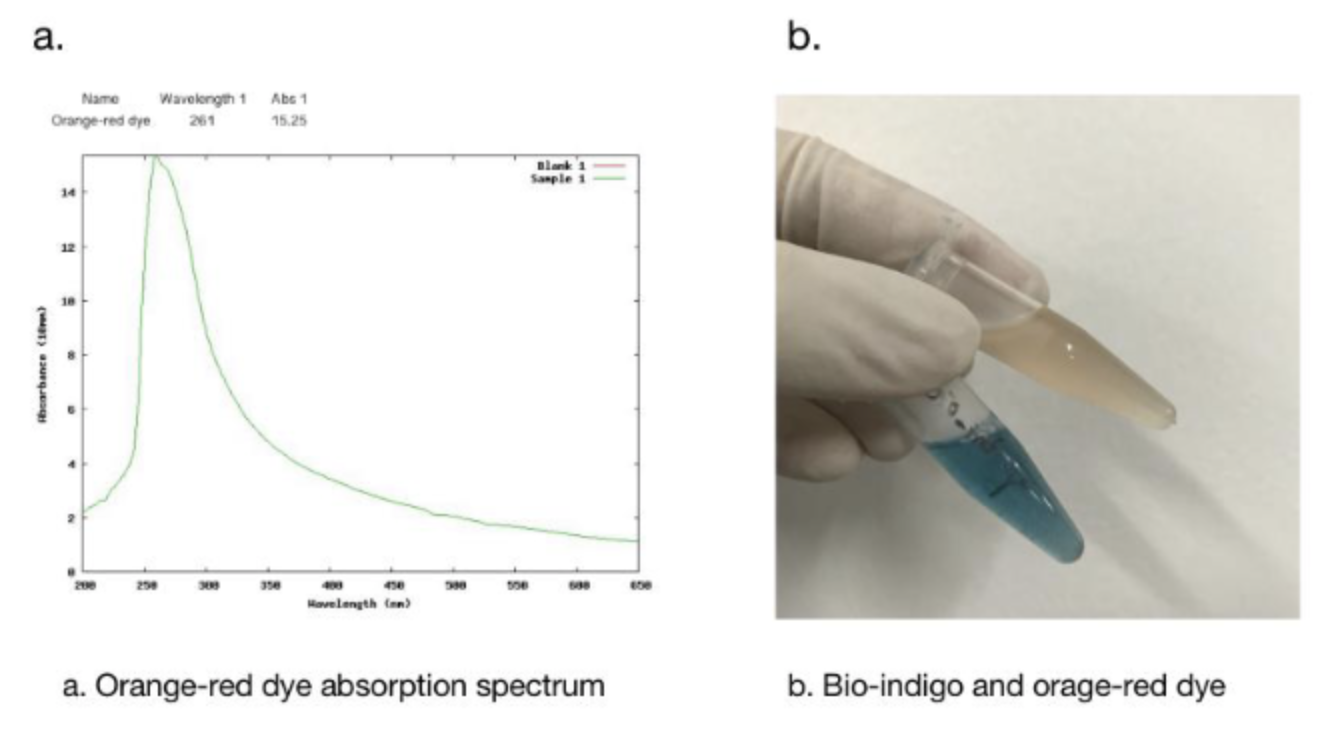
We used DMSO for resolubilization, and bio-indigo had an obvious absorption peak at 617nm, which was the same as that recorded in the literature. And the pigment is clearly blue in daylight.
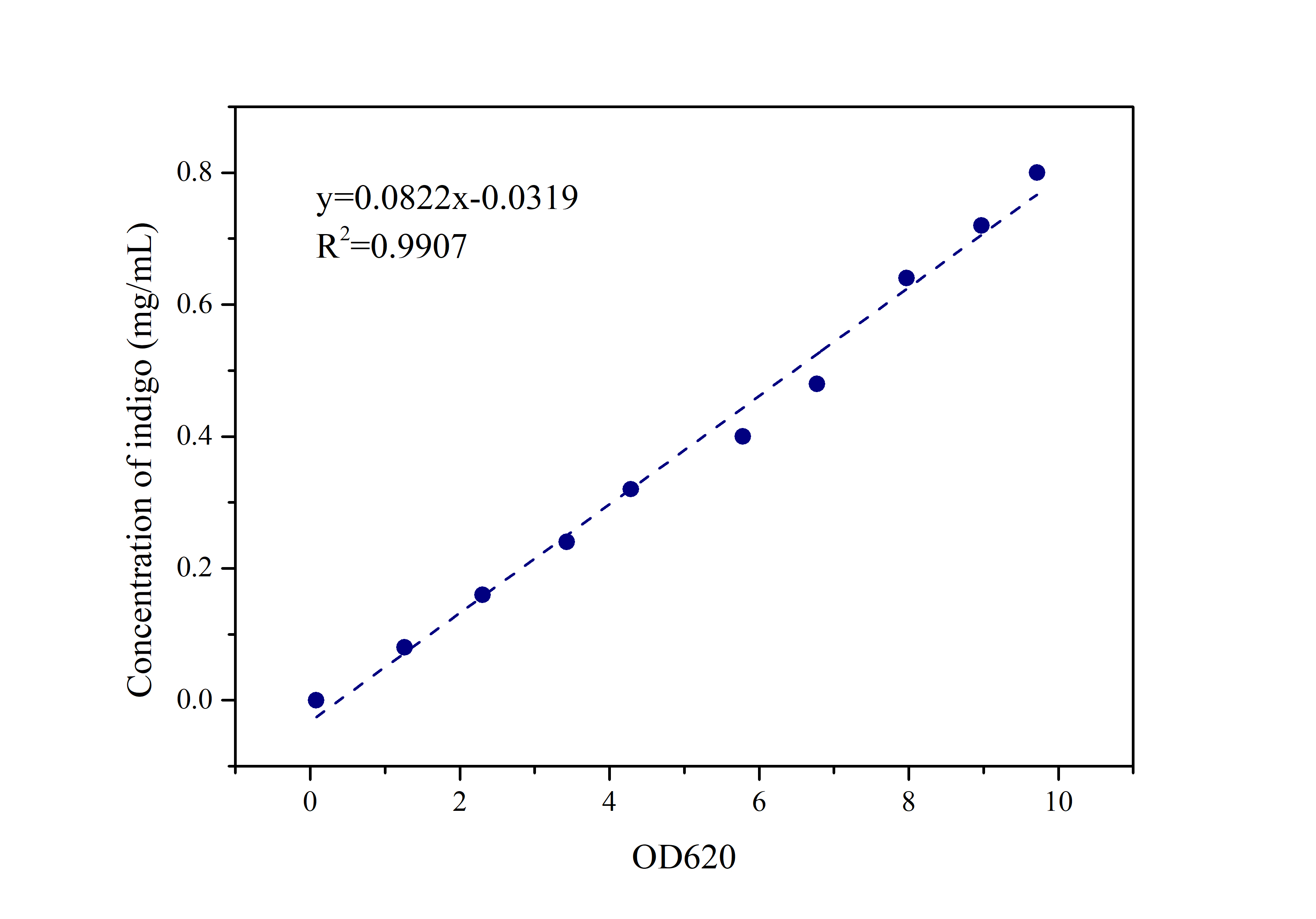
2.2 Indigo Standard Curve
Prepare indigo standard samples with sample concentrations of 0.8mg/ml, 0.72mg/ml, 0.64mg/ml, 0.48mg/ml, 0.4mg/ml 0.32mg/ml, 0.24mg/ml, 0.16mg/ml, 0.08mg /ml, 0mg/ml, measure the absorbance at 620 nm with an ultraviolet-visible spectrophotometer, and draw a standard curve.
2.3 Determination of Bio-indigo
ProductionThe cells were harvested by centrifugation at 110,000 rpm for 5 mins, after which the supernatant was removed and the precipitation was rinsed by distilled water twice.
Then, the cell pellet was resuspended by dimethyl sulfoxide (DMSO), and tested by ultraviolet-visible spectrophotometer UV-2250 for optical density at 620 nm.
Compare the data obtained in the experiment with the standard curve of the indigo sample to determine the indigo production.
In addition, we got the unexpected orange-red dye in the fermentation product. After we contacted with SHANGHAI_SFLS_SPBS, we found that this phenomenon also happened in their experiments. After consulting the literature, we found that this may be due to the production of isatin, which is obtained by oxidation of indigo. It is preliminarily speculated that this is due to the overexpression of FMO, which caused the indigo to be further oxidized into isatin.
3.Documentary Characterization
3.1 Background
FMO
Flavin-containing monoocygenases (FMOs) are first discovered in rat liver microsomes, and are usually found in eukaryotes. The amount of enzyme present in different tissues varies with species and sex, but the highest concentration is usually found in the liver. FMOs are involved in a wide range of oxidative biological processes, including drug detoxification, xenobiotic metabolism and bio-catalytic synthesis by catalyzation of the oxygenation of many nitrogen-, sulfur-, phosphorous-, selenium-, and other nucleophilic heteroatom.
In the literature in 2003, Choi, H.S.et alcloned the first discovered bacterial FMO from a methylotrophic bacterium they isolated from the seawater at Mokpo, Korea [1]. And they named the new species Methylophaga aminisulfidivorans SK1. It grows on methanol, methylated amines, and dimethylsulfide but not on methane. Fructose and glucose are the only multicarbon compounds tested that can be used as growth substrates.
They isolated and cloned a gene of Methylophaga aminisulfidivorans SK1 whose presence in E.coli produced blue dye indigo. The deduced amino acid sequence from the gene showed approximately 30% identities with FMOs of human (FMO1-FMO5). Its biochemical properties such as substrate specificities and absorption spectra were similar to the eukaryotic FMO families. Thus, Hack Sun Choi et al assigned the enzyme to be a bacterial FMO. And the nucleotide sequence of the bacterial fmo gene has been deposited in the GenBank database under Accession No. AF494423.
We used Clustal X to compare the nucleotide and amino acid sequence of FMOfrom the reference with that from BBa_1131000. We found that they have the same amino acid sequence, except for the redundant two amino acids, GS, on the C terminus of bFMO from BBa_1131000. But they have different DNA sequence with 77% identities.
Indigo
The use of indigo can date back to 6,000 years ago. It has been a prevalent dye contributing to the signature tone of blue denim. Indigo have a special way for dying. As indigo is insoluble, it have to be reduced to leuco-indigo for dying. And leuco-indigo can absorbs into fabric while diping and then rapidly oxidized to insoluble indigo after aeration. Thus, indigo can dye cotton without covalent bonds. Indigo as a dye has its own unique properties that’s irreplaceable. While absorbed indigo is robust to detergent for laundering contributing to durability, it is also susceptible to abrasion generating special fraying effect.
Historically, indigo was extracted form dye-producing plants, such as Indigofera sp.and Polygonum tinctorium. Due to the limited production of indigo extracted from plants, traditional extraction technology was rapidly replaced by the emergent chemical synthethic way in 19th. However, chemical synthetic way of indigo pose a serious threat to the environment, as it need toxic raw materials like aniline derived from petroleum, and hazardous chemical for processing, like reducing agent, formaldehyde, hydrogen cyanide, sodamide, and strong base.
Enduring high demand for indigo stimulate the emergence of the much more sustainable way for generating indigo, which appears to be synthesizing heterologous bio-indigo via fermentation.
3.2 bFMO Characteristics
3.2.1 Catalytic Mechanism of FMO
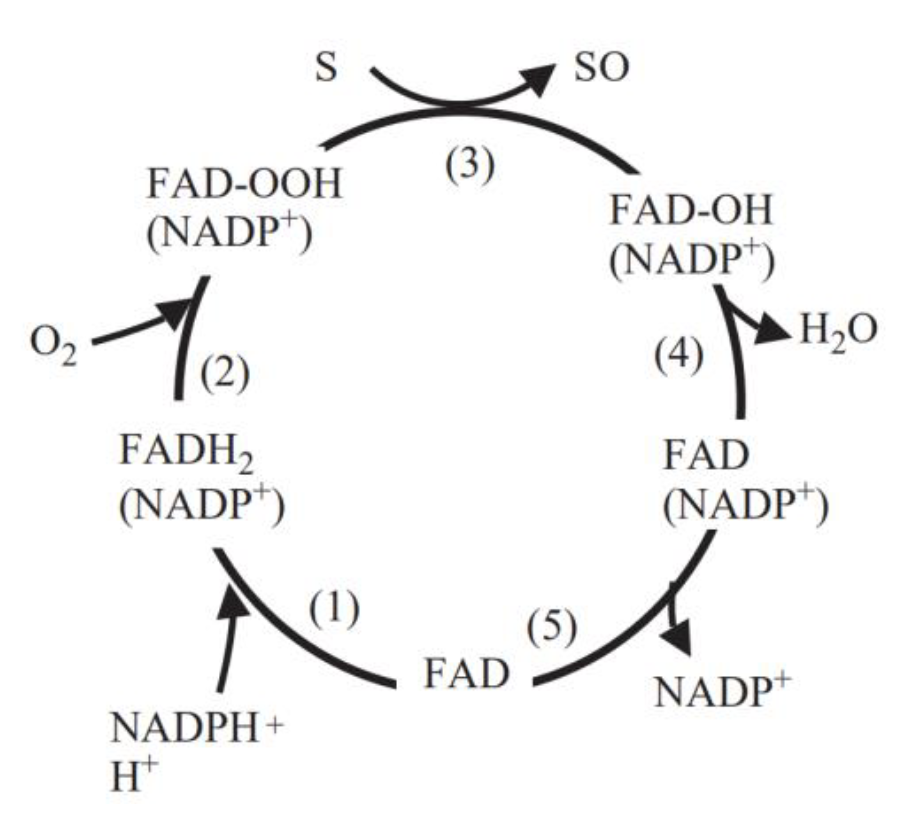
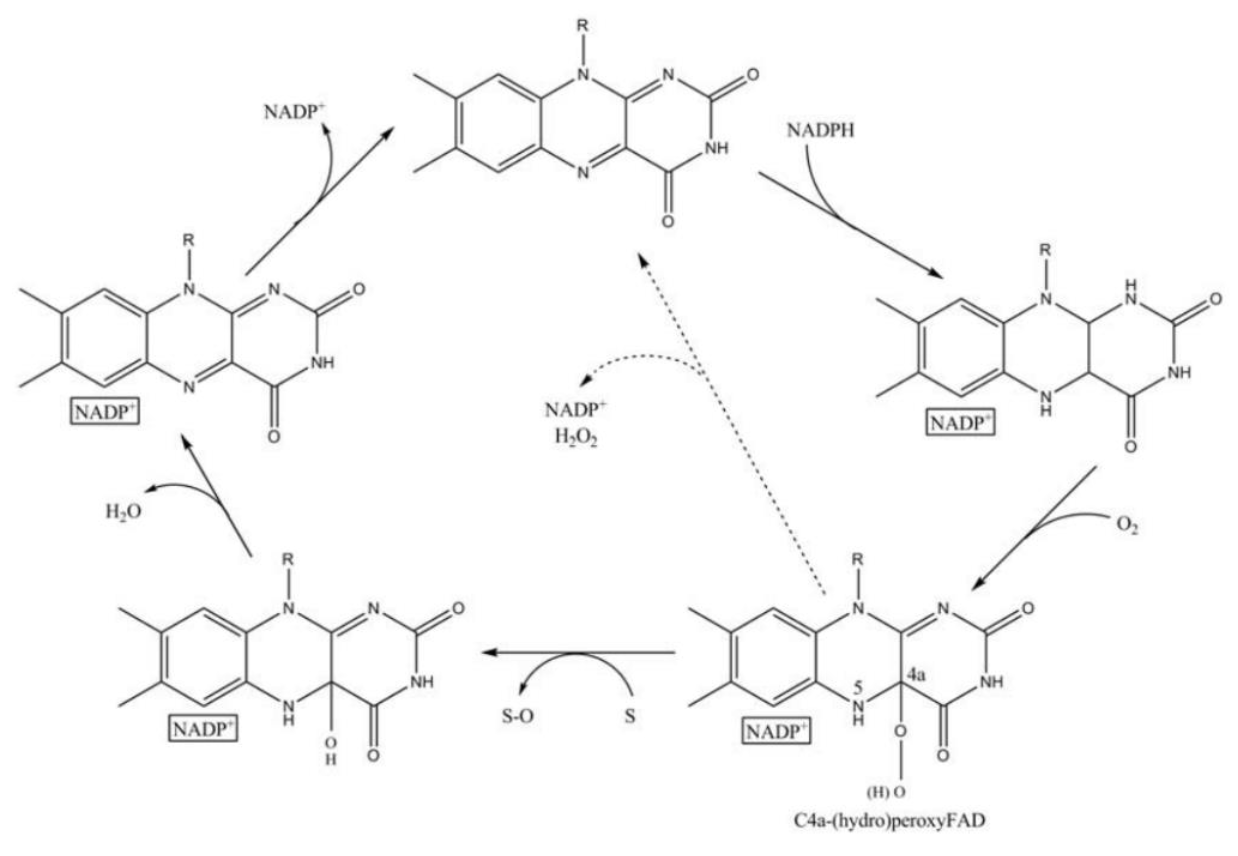
FMO belongs to a class of monooxygenases capable of generating a stable C4a peroxyflavin intermediate. In the first step of the catalytic cycle, FAD undergoes 2-electron reduction by NADPH (Fig. 5). The reduced flavin reacts rapidly with molecular oxygen to form the peroxyflavin and it is in this state, that FMO may exist predominantly in the cell, waiting for a suitable nucleophile with which to react. The nucleophilic attack by the substrate on the FADOOH results in 1 atom of molecular oxygen being transferred to the substrate and 1 atom to form water. The rate-limiting steps in the catalytic cycle are thought to be the breakdown of the FADOH psuedobase or the release of NADP+ . In either case, it is important to note that, unlike the CYP monooxygenase system, substrate binding has no influence on Vmax[2].
And the most special aspect in this catalytic cycle is that the presence of NADP+ is of critical importance to the stability of the intermediate, and it can keep binding to the enzyme before it finally release at the final step[3]. Binding of NADP can prevent enzyme from being oxidized by NADPH oxidase (Fig. 6).
3.2.2 Biochemical Characteristics of bFMO
In terms of substrate specificity, many substances can serve as good substrates of FMO, including trimethylamine, methiazole, nicotine, N, N-dimethylaniline and indoles, among which the substrate trimethylamine has the highest catalytic efficiency.
The gene encodingthe first discovered bacterial FMO was originally cloned from Methylophaga aminisulfidivorans SK1 [1], and was responsible for producing, the blue pigments, the blue indigo. The complete reading frame was1371bp long, which encodes a protein of 456 amino acids. The molecular mass of the encoded protein was 105 kDa, consisting of homodimer of 54kDa with an isoeletric point of 5.14. It contained 3characteristic sequence motifs of FMOs: FAD binding domain, FMO-identifying suquence motif, and NADPH binding domain.
FMOs of all mammals have strong binding to the cell membrane, and have low water solubility. Thus, up to now no mammalian FMO crystal structure has been obtained. Compared to eukaryotic FMOs, prokaryotic FMOs are more suitable for the construction of genetically engineered strains and can be better used in biocatalytic production. So its beyond dispute that bFMO has a promising future in bio-synthetic fiel.
3.2.3 Involvement in Bio-indigo Synthesis
Amodifiedsyntheticpathway for severalindigoidcompounds inthe recombinant E. coli containing FMO(AF494423)is representedin Fig. 7. In a tryptophan rich condition, extracellular tryptophanis transported into the cell by tryptophan permease and thereafterconverted into indole, pyruvate, and ammonia by tryptophanase(TnaA; EC4.1.99.1). FMO catalyzes thehydroxylation of indole to 2-hydroxyindole and 3-hydroxyindoleby using the reducing power of NADPH in the presence of oxygen. In the non-enzymatic reaction, indigo is producedfrom the combination of two 3-hydroxyindole molecules, whereasindirubin is made from the dimerization of 2-hydroxyindole and3-hydroxyindole[4].
3.3 Relevant Documentary Experiment
Optimization to Increase Yield of Indigo
After finding the gene encoding bFMO in Methylophaga aminisulfidivorans MPT, the members in the team of Chosun University, including Si Wouk Kim, keep moving on to optimize the condition of fermentation for increasing bio-indigo production by recombinant E.coli harboring fmo gene.
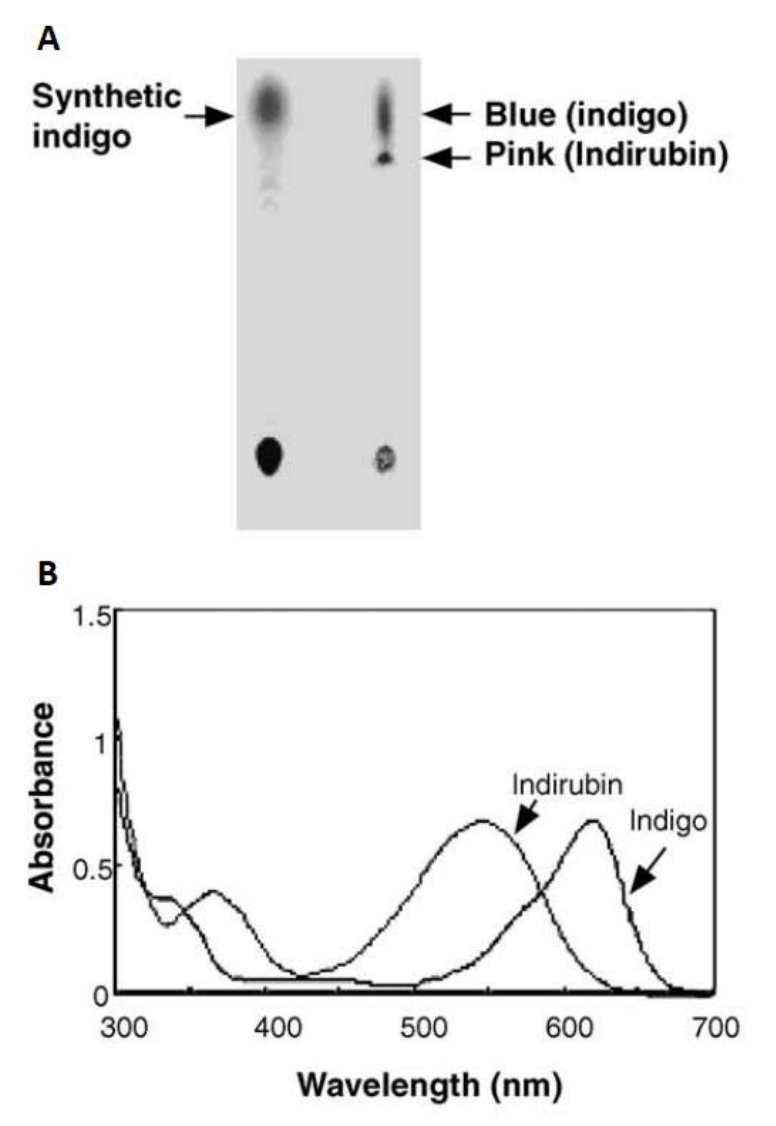
Initially in 2003, they constructed a plasmid pBlue 2.0 to express the fmo gene in E.coli, and achieved 160mg of indigo per liter in the trytophan medium (5 g yeast extract, 10 g NaCl, 2 g tryptophan per liter)supplementedwith ampicillin (50 lg/ml)after 12h cultivation at 37°C. The column purifiedblue pigments were analyzed by silica gel thin layer chromatography, and after migration the bacterial pigments separated into a predominant blue spot and a light pink spot (Fig. 8). To separate them, the mixture was injected into the HPLC (Waters) and fractionated by every 1 min at the wavelength of 620 nm. Two major active fractions were obtained with retention times of 8-11 and 15-18 min, respectively. The absorption spectra of blue and red fractions showed the highest absorption peaks at 620 and 540 nm (Fig. 8), corresponding to indigo and indirubin, respectively[1].
According to the literature of Si Wouk Kim et al in 2008, plasmid pBlue 2.0 consisted of a 2024 bp DNA fragments from Methylophaga aminisulfidivorans MPT, containing 651 bp upstream sequencesof fmo gene. To increase the production of indigo, they delete some of the upstream sequence and generate pBlue1.7 plasmid (1686bp) which proved to produce 575% increase in production of indigo, which is 920 mg per liter in optimum medium after 24h cultivation in fermentor. And the optimal medium was determined by using the response surface methodology, with tryptophan 2.4g/l, yeast extract 4.5 g/l and sodium chloride 11.4 g/l[5].
Protecting Group for Stabilizing Indoxyl
Even though microbial production of indigo can circumvents the use of harmful chemicals for synthesis, but a reducing agent is still required to reduce the insoluble indigo to the soluble form for dyeing.
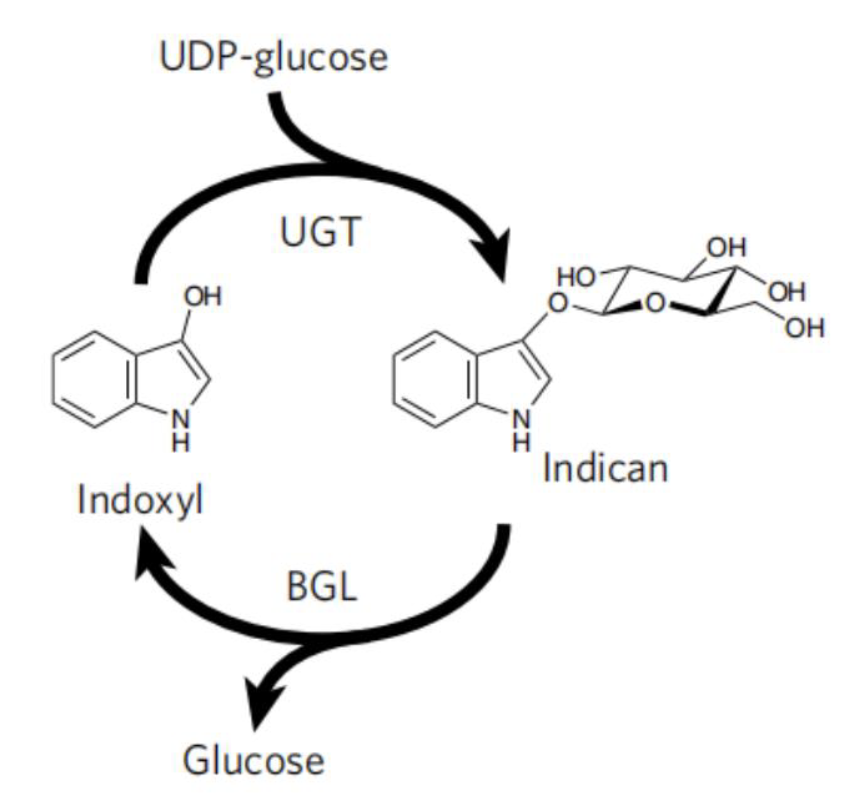
To avoid the use of reducing agent for dye solubilization, Tammy M.H. et al of University of California mimics the strategy employed by plant P.tinctorium[6]. This strategy utilize UDP-glycosyltransferase to glucosylates bio-synthetic indoxyl at the C3 hydroxyl group to form indican before its spontaneousair oxidation to indigo (Fig. 9). This glucoside, with glucose moiety as a biochemical protecting group is stable in air and can be stored. When dying, the protecting group canbe easily removed by using of beta-glucosidase, and generate indoxyl that can be oxidized to crystalline indigo in fibers after aeration. This brilliant strategy is a promising way for making up the deficiency in the sustainable scheme for bio-indigo production.
Reference
[1]Choi, H.S., et al., A novel flavin-containing monooxygenase from Methylophaga sp. strain SK1 and its indigo synthesis in Escherichia coli. Biochemical and Biophysical Research Communications, 2003. 306(4): p. 930-936.
[2]Krueger, S.K. and D.E. Williams, Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther, 2005. 106(3): p. 357-87.
[3]Zhenxuan, W., Cloning, expression and the catalytic mechanism research of a bacterial flavin-containing monooxygenase. Shanghai Jiao Tong University, 2011.
[4]Han, G.H., et al., Enhanced indirubin production in recombinant Escherichia coli harboring a flavin-containing monooxygenase gene by cysteine supplementation. J Biotechnol, 2012. 164(2): p. 179-87.
[5]Han, G.H., H.-J. Shin, and S.W. Kim, Optimization of bio-indigo production by recombinant E. coli harboring fmo gene. Enzyme and Microbial Technology, 2008. 42(7): p. 617-623.
[6]Hsu, T. M.et al., Employing a biochemical protecting group for a sustainable indigo dyeing strategy. Nat Chem Biol, 2018. 14(3):p. 256-261
==BJU-China 2021 iGEM TEAM==
TnaA is endogenous tryptophanase from E. coli that can convert tryptophan to indole. MaFMO is monooxygenase from Methylophaga aminisulfidivorans which could effectively synthesize indigo by oxidation of indole.
Description
In our project, TnaA-MaFMO is a combination used in 6BrIG production strategy in E. coli BL21(DE3). Since E. coli inherently carries TnaA, to prevent the influence of endogenous expression, we knock out TnaA in the genome, and then compensate TanA on the pET28b plasmid together with MaFMO. The expression of TnaA-MaFMO was controlled by T7-LacO inducible promoter, and IPTG acts as inducer.
Experiments and Results
We obtained TnaA gene and MaFMO gene by PCR and then assembled them to pET28b backbone by Gibson. Colony PCR and gene sequencing were use to verify the sequences. The following was the agarose gel electrophoresis of the results (Figure 2).
Next, we want to test the expression of these enzymes with IPTG induction. For better expression of enzymes, we optimize the induction condition of gene expression: three different temperatures (18℃, 30℃, 37℃) and inducer concentrations (0mM, 0.1mM, 1mM) were set for test. We successfully detected TnaA-MaFMO enzyme after IPTG induction using SDS-PAGE (Figure 3). In the gel picture, we can clearly see TanA-MaFMO bands (about 52.8 KDa). And we can find that TanA-MaFMO expresses significantly better at 30℃ and 37℃ than 18℃. When the inducer concentration is lower (0.1mM), the protein bands are more obvious, and even when there is no inducer, the gene also have a basic expression, that is also called leakage. The leakage may caused by high gene expression using high copy plasmid pET28b.
In summary, we successfully constructed a plasmid including two enzymes TnaA and MaFMO, and detected their expression through the induction of IPTG.