Difference between revisions of "Part:BBa K3771092"
Karin100699 (Talk | contribs) |
Karin100699 (Talk | contribs) |
||
| Line 17: | Line 17: | ||
<br> | <br> | ||
| − | <br>This composite part was ligated with the pSAA vector and transformed into <i>E. coli</i>. We conducted colony PCR to verify whether <i>E. coli</i> uptake the correct plasmid. The size of the PCR product was as expected.The part has been confirmed by sequencing and has no mutations. | + | <br>This composite part was ligated with the pSAA vector and transformed into <i>E. coli</i>. We conducted colony PCR to verify whether <i>E. coli</i> uptake the correct plasmid. The size of the PCR product was as expected. The part has been confirmed by sequencing and has no mutations. |
<br> | <br> | ||
Revision as of 12:21, 18 October 2021
PsoxS-sfGFP
Description
This composite part consists of promoter PsoxS (BBa_K3771048) and sfGFP (BBa_K1321337). The soxS gene is one of the soxRS regulon components, which is important for E. coli to sense and respond to the oxidants. The soxS promoter (PsoxS) is stimulated by oxidative stress, leading to the expression of the downstream gene. sfGFP serves as a reporter to evaluate the strength of PsoxS.
Usage and Biology
This composite part was ligated with the pSAA vector and transformed into E. coli. We conducted colony PCR to verify whether E. coli uptake the correct plasmid. The size of the PCR product was as expected. The part has been confirmed by sequencing and has no mutations.

Fig. 1. (A) The electrophoresis result of colony PCR. M: A.Marker; Lane 1: pSAA-J23100-sfgfp (2537 bp); Lane 2, 3: pSAA-PsoxS-sfgfp (3122 bp). (B) Digested the amplified fragments with HindIII. M: Marker; Lane 1-3: pSAA-PsoxS-sfgfp (378, 396, 1933 bp); Lane 4: pSAA-J23100-sfgfp.
We conducted a disk assay to examine what kinds of oxidative stress inducers PsoxS can react to. First, 0.5ml of overnight culture was mixed with 3ml of melted 0.5% soft agar LB at 50˚C and overlaid on a LB plate. Next, we put paper discs on the plate and dropped solutions to induce oxidative stress on paper discs. Last, we incubated the plate at 37˚C for 27 hours and checked the result. In our result, PsoxS responds to both low- and high-concentration hydrogen peroxide. It also responds to menadione weakly and responds to paraquat effectively (Fig. 2).

Fig. 2. The disk assay result of PsoxS responding to different oxidative stress inducers. H2O2s - 30% hydrogen peroxide, 10ul; H2O2w - 3% hydrogen peroxide, 10ul; MD - 10mM menadione, 10ul; DMSO - solvent for menadione, 10ul; PQ - 1mM paraquat, 10ul; MQ - solvent for paraquat, 10ul.
The promoter strength of PsoxS is determined by the expression level of the reporter, sfGFP, under oxidative stress. While using hydrogen peroxide (H2O2) as the inducer, the intensity of PsoxS shows no significant difference under different concentrations of hydrogen peroxide (Fig. 3). However, PsoxS is well induced by using paraquat (PQ), which is a commonly used agent to induce oxidative stress for bacteria (Fig. 4 & 5).

Fig. 3. Relative fluorescence intensity of PsoxS after 4.5-hour incubation with hydrogen peroxide in various concentrations.
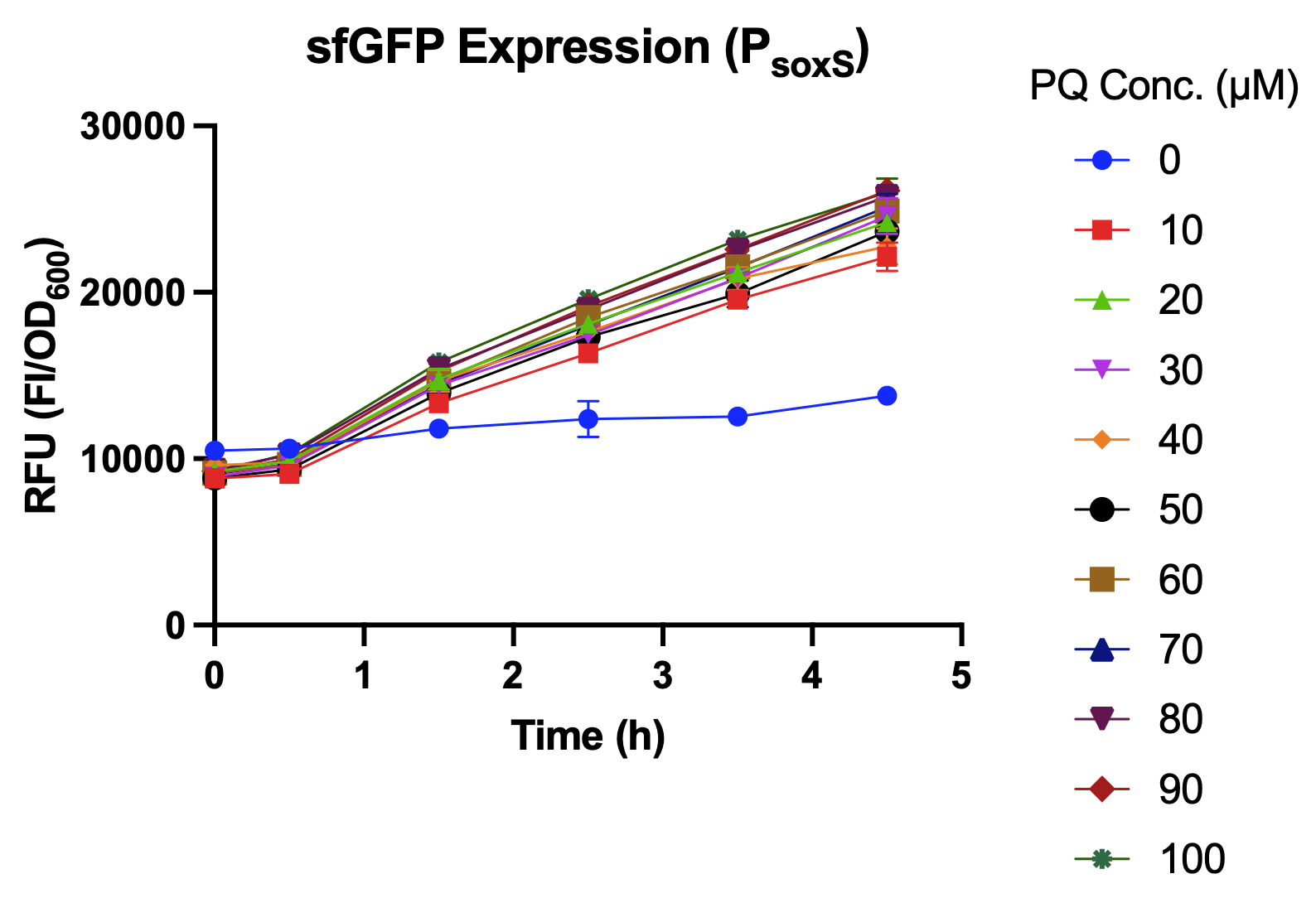
Fig. 4. Relative fluorescence intensity of PsoxS after 4.5-hour incubation with hydrogen peroxide in low concentrations.

Fig. 5. Relative fluorescence intensity of PsoxS after 4.5-hour incubation with hydrogen peroxide in high concentrations.
We compared the relative fluorescence intensity of sfGFP expressed by distinct promoters under the stimulation by different concentrations of paraquat. As expected, sfGFP expression level of J23100 shows no difference between the groups with and without paraquat. PsoxS with its activator, SoxR (BBa_K37710XX), is highly activated under oxidative stress and has the highest gene expression level compared to J23100, PkatG (BBa_K37710XX), and PsoxS (BBa_K37710XX) solely (Fig. 6). Moreover, we compared the efficiency of our design with (BBa_K37710XX), in which the concept came from the 2018 Imperial College London iGEM team. Different from our design, they used a mutant PsoxS promoter with RiboJ (BBa_K2862010) and expressed SoxR by J23101 promoter. However, the sfGFP expression by BBa_K2862010 was quite low (Fig. 7).

Fig. 6. Comparison of different promoter strengths by relative fluorescence intensity after 4.5-hour incubation with paraquat in high concentrations.

Fig.7. Comparison of different promoter strengths by relative fluorescence intensity after 4.5-hour incubation with paraquat in low concentrations.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 123
