Difference between revisions of "Part:BBa R0187"
JackSuitor (Talk | contribs) |
|||
| Line 5: | Line 5: | ||
A lac repressible T7 promoter. This promoter is similar to those found in the pET vectors with T7lac promoters. I mutated the -9 base to an G, which should make it less than 3% of the strength of consensus. The base numbering is as used by Studier and coworkers (JMB 1991, 219, 45-59). See BBa_R0183 for more information. | A lac repressible T7 promoter. This promoter is similar to those found in the pET vectors with T7lac promoters. I mutated the -9 base to an G, which should make it less than 3% of the strength of consensus. The base numbering is as used by Studier and coworkers (JMB 1991, 219, 45-59). See BBa_R0183 for more information. | ||
Note: This part has been improved by Edinburgh_UG 2017. See their improved part here: <partinfo>BBa_K2406020</partinfo> | Note: This part has been improved by Edinburgh_UG 2017. See their improved part here: <partinfo>BBa_K2406020</partinfo> | ||
| + | |||
| + | =SHSBNU_China 2021= | ||
| + | In this project, we used combined part of T7 and LacI binding site (BBa_R0187) in our gene circuit. This part is the regulatory region in our circuit that is responsible for the controlled expression of our proteins (BBa_K3798015, BBa_K3798033) that we have experimented with different induction intensity to obtain the best IPTG usage. | ||
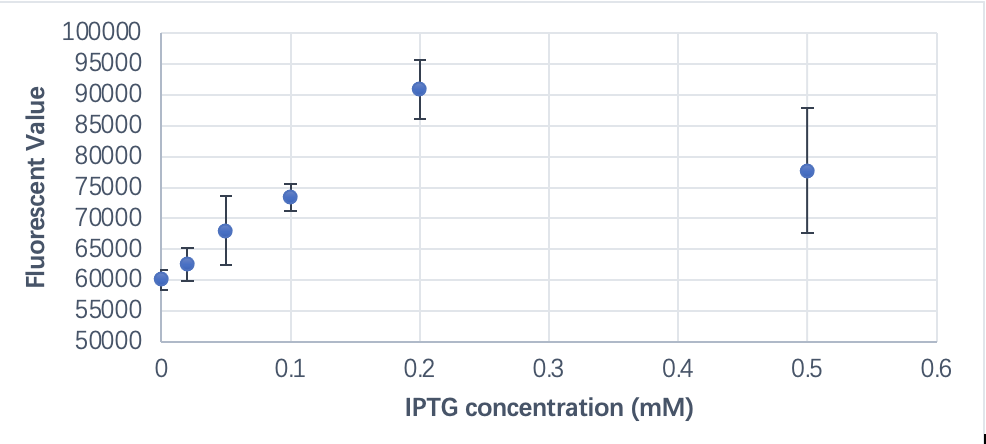
| + | To express sufficient amount of CBD-eGFP protein for our use, we applied different concentrations of IPTG to the induction process of e.coli incubation. | ||
| + | [[File:T--SHSBNU_China--Contribution 1.png|600px|thumb|center]] | ||
| + | <p class="figure-description"><b><center>Fig.1 The relationship between fluorescent value and IPTG concentration</center></b></p > | ||
| + | As shown in the chart, the fluorescent value increases with the increased IPTG concentration. Although in the diagram the leakage of this T7 promoter seems significant, this is actually caused by the use of LB medium in our culture that interfered with the fluorescent value. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 12:43, 14 October 2021
T7 promoter (lacI repressible)
A lac repressible T7 promoter. This promoter is similar to those found in the pET vectors with T7lac promoters. I mutated the -9 base to an G, which should make it less than 3% of the strength of consensus. The base numbering is as used by Studier and coworkers (JMB 1991, 219, 45-59). See BBa_R0183 for more information. Note: This part has been improved by Edinburgh_UG 2017. See their improved part here: BBa_K2406020
SHSBNU_China 2021
In this project, we used combined part of T7 and LacI binding site (BBa_R0187) in our gene circuit. This part is the regulatory region in our circuit that is responsible for the controlled expression of our proteins (BBa_K3798015, BBa_K3798033) that we have experimented with different induction intensity to obtain the best IPTG usage. To express sufficient amount of CBD-eGFP protein for our use, we applied different concentrations of IPTG to the induction process of e.coli incubation.
As shown in the chart, the fluorescent value increases with the increased IPTG concentration. Although in the diagram the leakage of this T7 promoter seems significant, this is actually caused by the use of LB medium in our culture that interfered with the fluorescent value.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]