Difference between revisions of "Part:BBa K112809"
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K112809 short</partinfo> | <partinfo>BBa_K112809 short</partinfo> | ||
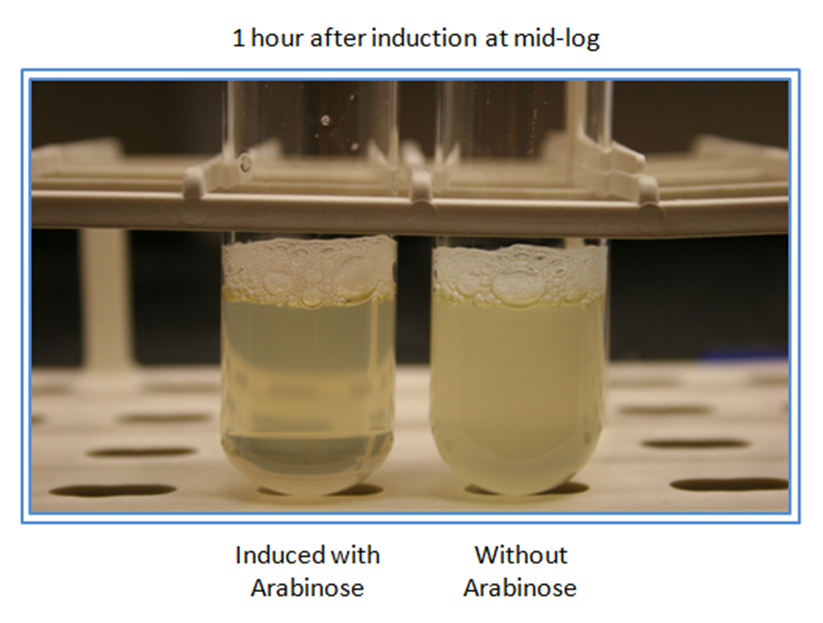
This is the T4 Lysis Device with Pbad. Adding arabinose at mid-log lyses the cell very well. It also lyses cells at saturation if it is on a high copy plasmid. | This is the T4 Lysis Device with Pbad. Adding arabinose at mid-log lyses the cell very well. It also lyses cells at saturation if it is on a high copy plasmid. | ||
| + | |||
| + | ==Characterization== | ||
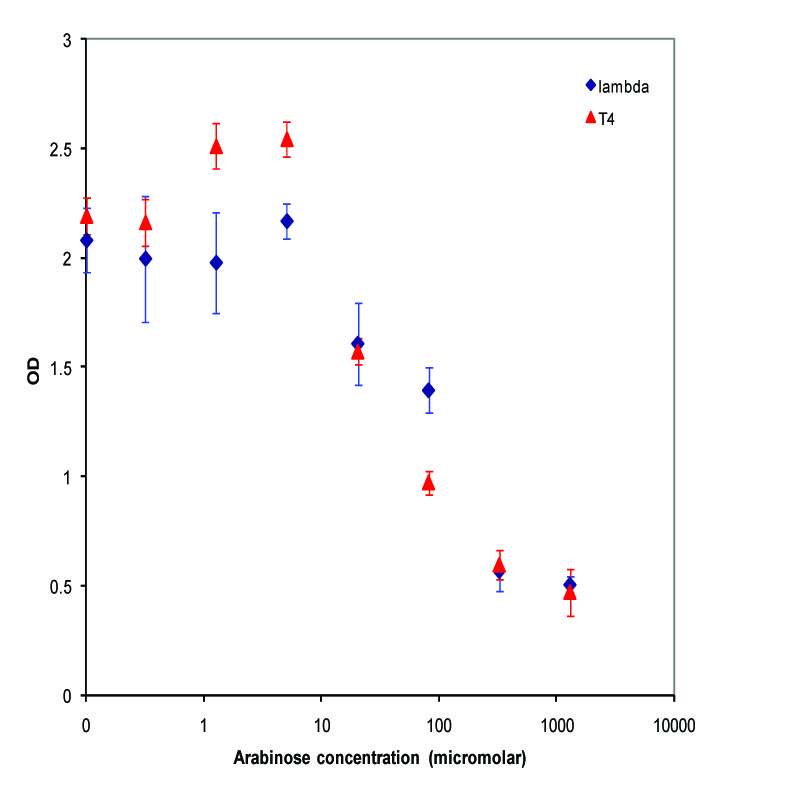
| + | K112019 (Lambda Lysis Device) was inserted in the vector K112950 and K112809 was insertedin pSB1A2. They were introduced into the MC1061 strain by transformation, and then picked 5 colonies for each device. We grew these cultures to saturation at 37 degrees Celsius in LB media, and then split into eight 1 mL aliquots. A range of concentrations of arabinose was added to these aliquots, with a starting concentration of 1.3E-3 M and the next 6 samples recieving a four-fold dilution of the previous sample, and an equal volume of water added to the last aliquot. The cultures were then incubated at 37 degrees again for 3.5 hours, and the absorbance at 600nm was measured with a Tecan Xfluor4 Safire2 in a Corning Inc. Costar 3603 plate. The data plotted on a log scale is shown below. | ||
| + | |||
| + | |||
| + | [[Image:K112019_data1.jpg|thumbnail|450px|center|Data from adding arabinose at midlog. Concentrations used at each data point from left to right are 0, 2.44E-07, 9.77E-07, 3.91E-06, 1.56E-05, 6.25E-05, 2.50E-04, and 1.00E-03 M]] | ||
| + | |||
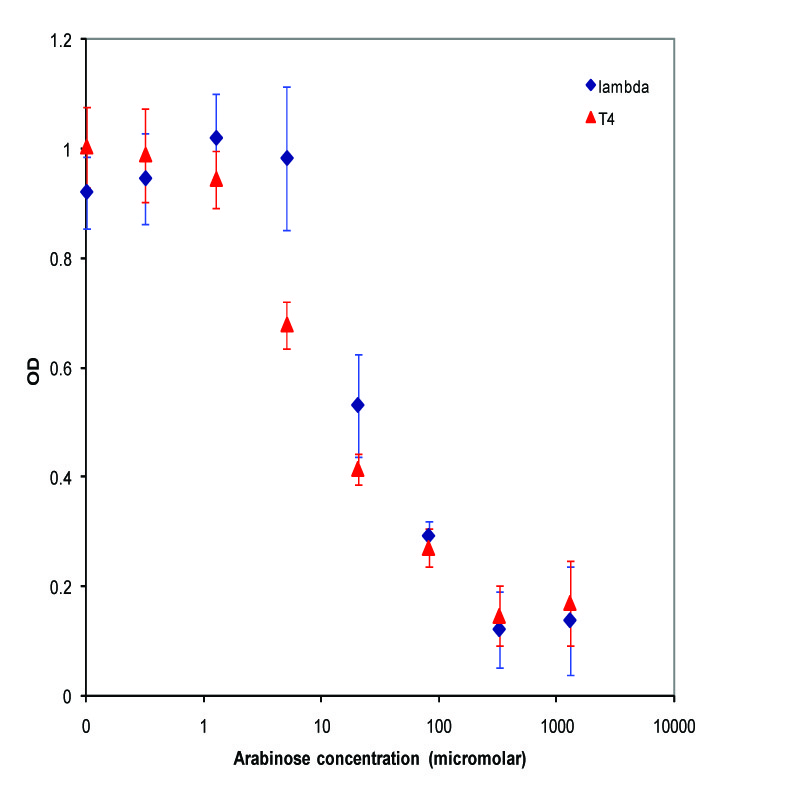
| + | A parallel experiment was performed by taking the same saturated culture before induction with arabinose, diluting 100-fold, growing to mid-log(starting OD .22), before inducing with arabinose as above. The data plotted on a log scale is shown below. | ||
| + | |||
| + | [[Image:K112019_data2.jpg|thumbnail|450px|center|Data from adding arabinose at midlog. Concentrations used at each data point from left to right are 0, 2.44E-07, 9.77E-07, 3.91E-06, 1.56E-05, 6.25E-05, 2.50E-04, and 1.00E-03 M]] | ||
| + | |||
| + | [[Image:lysis.jpg]] | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 08:22, 6 November 2008
T4 Lysis Device with Pbad
This is the T4 Lysis Device with Pbad. Adding arabinose at mid-log lyses the cell very well. It also lyses cells at saturation if it is on a high copy plasmid.
Characterization
K112019 (Lambda Lysis Device) was inserted in the vector K112950 and K112809 was insertedin pSB1A2. They were introduced into the MC1061 strain by transformation, and then picked 5 colonies for each device. We grew these cultures to saturation at 37 degrees Celsius in LB media, and then split into eight 1 mL aliquots. A range of concentrations of arabinose was added to these aliquots, with a starting concentration of 1.3E-3 M and the next 6 samples recieving a four-fold dilution of the previous sample, and an equal volume of water added to the last aliquot. The cultures were then incubated at 37 degrees again for 3.5 hours, and the absorbance at 600nm was measured with a Tecan Xfluor4 Safire2 in a Corning Inc. Costar 3603 plate. The data plotted on a log scale is shown below.
A parallel experiment was performed by taking the same saturated culture before induction with arabinose, diluting 100-fold, growing to mid-log(starting OD .22), before inducing with arabinose as above. The data plotted on a log scale is shown below.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1205
Illegal NheI site found at 2568
Illegal NheI site found at 2591 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1144
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
Illegal AgeI site found at 2173
Illegal AgeI site found at 2243 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961
Illegal SapI site found at 2824