Difference between revisions of "Part:BBa T9150"
| Line 6: | Line 6: | ||
Coding region for PyrF gene. PyrF is used in the synthesis of uracil in the same pathway that Ura3 is used in yeast. Can also be used as a conterselectable marker in the presence of 5-Fluoroorotic Acid (5-FOA) and the absence of uracil. | Coding region for PyrF gene. PyrF is used in the synthesis of uracil in the same pathway that Ura3 is used in yeast. Can also be used as a conterselectable marker in the presence of 5-Fluoroorotic Acid (5-FOA) and the absence of uracil. | ||
| − | + | ===Information contributed by City of London UK (2021)=== | |
| − | === | + | Part information is collated here to help future users of the BioBrick registry. |
| + | |||
| + | Metadata: | ||
| + | *'''Group:''' City of London UK 2021 | ||
| + | *'''Author:''' Lucas Ng | ||
| + | *'''Summary:''' Added information collated from existing scientific studies | ||
| + | |||
| + | ---- | ||
| + | |||
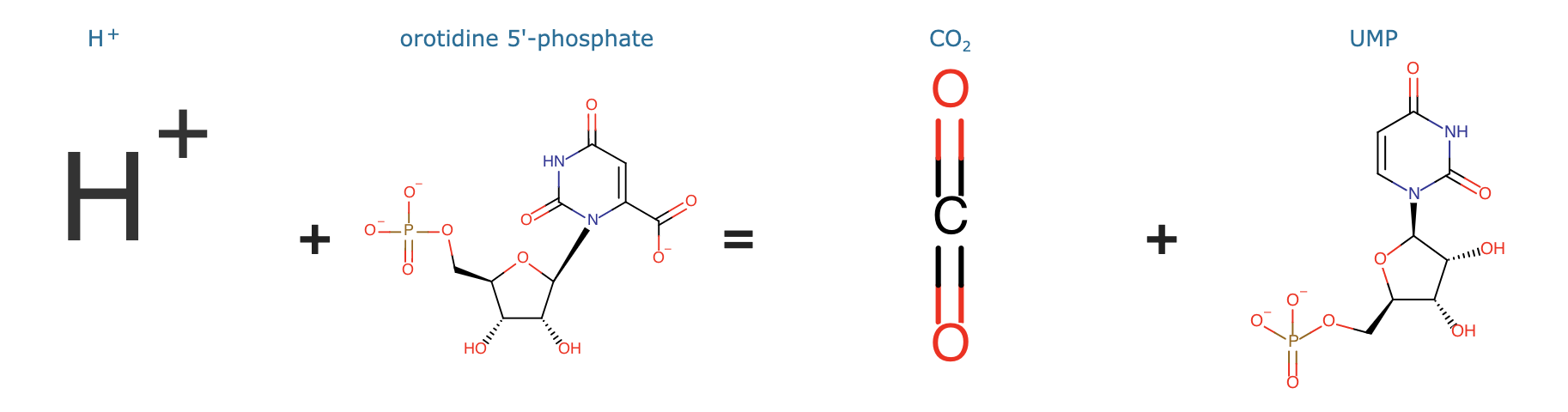
| + | PyrF codes for orotidine 5'-phosphate decarboxylase, which catalyzes the decarboxylation of orotidine 5'-monophosphate (OMP) to uridine 5'-monophosphate (UMP) | ||
| + | <ref>“Rhea - Annotated Reactions Database.” n.d. Rhea. Accessed July 27, 2021. https://www.rhea-db.org/rhea/11596. | ||
| + | </ref>: | ||
| − | + | [[File:PyrF_reaction.png|x200px|center]] | |
| − | + | ||
| − | + | ||
| − | + | Involved in pyrimidine metabolism, PyrF is part of the pathway biosynthesizing UMP from orotate <i>de novo</i> | |
| − | < | + | <ref> |
| − | === | + | Jensen, K. F., J. N. Larsen, L. Schack, and A. Sivertsen. 1984. “Studies on the Structure and Expression of Escherichia Coli PyrC, PyrD, and PyrF Using the Cloned Genes.” European Journal of Biochemistry 140 (2): 343–52. https://doi.org/10.1111/j.1432-1033.1984.tb08107.x. |
| − | + | </ref>. | |
| − | + | ||
| + | ===References=== | ||
Latest revision as of 16:31, 27 July 2021
orotidine 5
Coding region for PyrF gene. PyrF is used in the synthesis of uracil in the same pathway that Ura3 is used in yeast. Can also be used as a conterselectable marker in the presence of 5-Fluoroorotic Acid (5-FOA) and the absence of uracil.
Information contributed by City of London UK (2021)
Part information is collated here to help future users of the BioBrick registry.
Metadata:
- Group: City of London UK 2021
- Author: Lucas Ng
- Summary: Added information collated from existing scientific studies
PyrF codes for orotidine 5'-phosphate decarboxylase, which catalyzes the decarboxylation of orotidine 5'-monophosphate (OMP) to uridine 5'-monophosphate (UMP) [1]:
Involved in pyrimidine metabolism, PyrF is part of the pathway biosynthesizing UMP from orotate de novo [2].
References
- ↑ “Rhea - Annotated Reactions Database.” n.d. Rhea. Accessed July 27, 2021. https://www.rhea-db.org/rhea/11596.
- ↑ Jensen, K. F., J. N. Larsen, L. Schack, and A. Sivertsen. 1984. “Studies on the Structure and Expression of Escherichia Coli PyrC, PyrD, and PyrF Using the Cloned Genes.” European Journal of Biochemistry 140 (2): 343–52. https://doi.org/10.1111/j.1432-1033.1984.tb08107.x.