Difference between revisions of "Part:BBa K3410001"
| Line 33: | Line 33: | ||
Error Prone PCR | Error Prone PCR | ||
For means of affinity maturation we needed a library of our grafted sequence. To introduce the required diversity into the grafted sequence, we chose a random mutagenesis approach by polymerase chain reaction (PCR), first described by Cadwell and Joyce in 1992 [28]. This variant of the PCR-technique is termed error prone PCR (epPCR). In this technique, the natural error rate of the Taq polymerase is increased by unfavorable reaction conditions. This is accomplished by: 1) Increased concentration of Taq DNA polymerase; 2) extended polymerase extension time; 3) increased concentration of MgCl2 ions; 4) unbalanced rate of dNTPs; and 5) addition of MnCl2 ions [29]. | For means of affinity maturation we needed a library of our grafted sequence. To introduce the required diversity into the grafted sequence, we chose a random mutagenesis approach by polymerase chain reaction (PCR), first described by Cadwell and Joyce in 1992 [28]. This variant of the PCR-technique is termed error prone PCR (epPCR). In this technique, the natural error rate of the Taq polymerase is increased by unfavorable reaction conditions. This is accomplished by: 1) Increased concentration of Taq DNA polymerase; 2) extended polymerase extension time; 3) increased concentration of MgCl2 ions; 4) unbalanced rate of dNTPs; and 5) addition of MnCl2 ions [29]. | ||
| + | But how does grafting of antibodies work? A quick guide how to graft: | ||
| + | |||
| + | In a first step of in silico nanobody grafting, the amino acid sequences of both donor (10G6D6 and C12G11, respectively) and acceptor (3DWT) are numbered. There are different numbering schemes for immunoglobulin variable domains available [9], we decided to use the AHo numbering scheme, because placement of gaps is based on the spatial alignment of known three dimensional structures of immunoglobulin domains and not on sequence variability [10]. The numbering was performed using ANARCI (SAbPred) (http://opig.stats.ox.ac.uk/webapps/newsabdab/sabpred/anarci/). For further work, the numbered residues were transferred to an excel sheet. Both, the CDRs and the functional important framework residues of the donor (10G6D6 and C12G11) were combined with the acceptor (3DWT) and a new nanobody was created (Graft). | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild4.jpg|thumb|800px|center|Figure 4: Map of the grafted estradiol nanobody. The CDRs were taken from the mAb anti-estradiol antibody 10G6D6 and were subsequently grafted onto the scaffold – the nanobody 3DWT. For immobilisiation, a his tag was chosen and added. In accordance with our cloning strategy SfiI recognition sites were added at 5´ and 3´ ends as well as a 6 nt long appendix, so SfiI does not have to cut directly at the 5´ nor the 3´ end.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild5.jpg|thumb|800px|center|Figure 5: Map of the grafted progesterone nanobody. The CDRs were taken from the mAb anti-progesterone antibody 15G12C12G11 and were subsequently grafted onto the scaffold – the nanobody 3DWT. For immobilisiation, a his tag was chosen and added. In accordance with our cloning strategy, SfiI recognition sites were added at 5´ and 3´ ends as well as a 6 nt long appendix, so SfiI does not have to cut directly at the 5´ nor the 3´-end.]] | ||
| + | |||
| + | The grafting results with the exact sequence can be found in table 1 for our estradiol nanobody and in table 2 for our progesterone nanobody. | ||
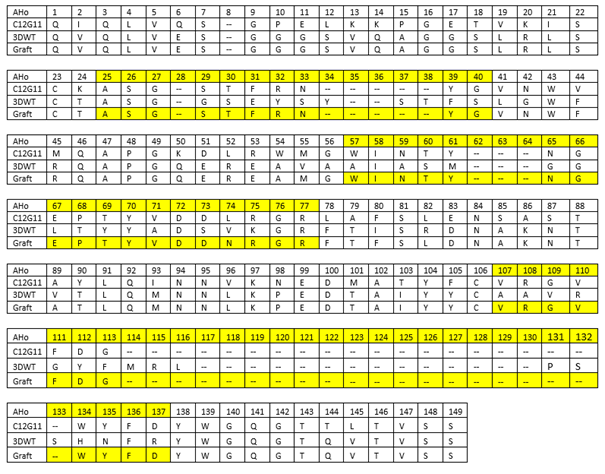
| + | Table 1: Design of the estradiol nanobody. The first 44 amino acid residues (AHo numbering) of the donor antibody (10G6), the accepting antibody (3DWT) and the grafted estradiol nanobody are shown. The complementarity determining regions are highlighted in yellow. | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild6.jpg|thumb|800px|center|Figure 6: Design of the progesterone nanobody. The first 44 amino acid residues (AHo numbering) of the donor antibody (C12G11), the accepting antibody (3DWT) and the grafted progesterone nanobody are shown. The complementarity determining regions are highlighted in yellow. | ||
| + | |||
| + | |||
| + | Using ChimeraX (https://www.cgl.ucsf.edu/chimerax/), we optically verified the 3D structure of the grafted nanobodies for steric obstacles to antigen binding: For example, we checked, if the grafted residues sterically impaired residues from the framework. | ||
| + | |||
| + | |||
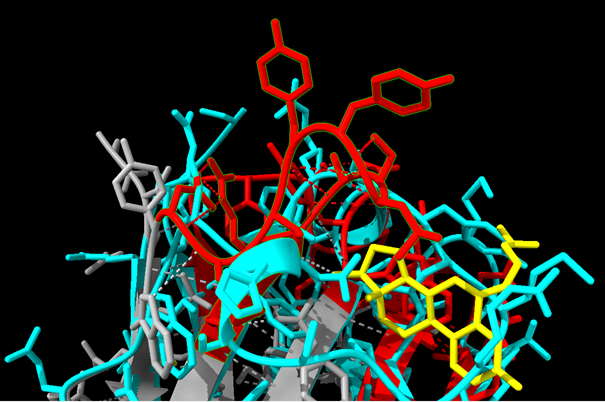
| + | [[File:T--Bielefeld-CeBiTec--Bild7.jpg|thumb|800px|center|Figure 7: Screenshot taken from ChimeraX analysing the estradiol graft. An alignment between the scaffold 3DWT and CDR donor (10G6D6) was performed to check for sterical impairments with the amino acid residues. Estradiol is depicted in yellow, the scaffold 3DWT in cyan and 10G6D6 is marked in grey. Furthermore, the three to be grafted CDRs from 10G6D6 are shown in red. Searching for impairments, we checked every amino acid residue for sterical clashes between the grafted amino acids and the remaining residues from the scaffold which are not neutralized by the grafting process.]] | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild8.jpg|thumb|800px|center|Figure 8: Screenshot taken from ChimeraX analysing the progesterone graft. An alignment between the scaffold 3DWT and CDR donor 15G12C12G11 was performed to check for sterical impairments with the amino acid residues. Depicted in white is progesterone. Highlighted in blue is the scaffold 3DWT and in red 15G12C12G11. Furthermore, shown in green are the three to be grafted CDRs from 15G12C12G11. Searching for impairments, we checked every amino acid residue for sterical clashes between the grafted amino acids and the remaining residues from the scaffold which are not neutralized by the grafting process.]] | ||
| + | |||
| + | Subsequently, the amino acid sequences were translated into a complementary DNA sequence. This was performed using the tool Sequence Manipulation Suite (http://bioinformatics.org/sms2/rev_trans.html). | ||
| + | |||
| + | Furthermore, the sequences were optimized in regard of their GC content and number of repeats as well as their length. The created nucleotide sequences were purchased as gene synthesis from IDT and EurofinsGenomics. In general, the nanobody grafting process generates weak binders as the sequence of the scaffold is strongly modified, especially with regard to the CDRs, which can partly form new secondary structures. In addition, core frame work residues are also transferred from the donor to the scaffold, which changes its sequence, perhaps resulting in a changed structure of the scaffold in non-CDR areas. Therefore, the grafted nanobodies have to be improved by suitable affinity maturation measures [9]. | ||
| + | |||
| + | |||
| + | Because the epPCR conducted with the OneTaq® DNA Polymerase showed better results regarding positive clones (gelbild colony PCR (see Fig. 6) as well as more and larger red colonies after transformation (see Fig. 3) compared to the results of the Taq polymerase, we decided to use OneTaq® DNA Polymerase for our epPCR experiments using the protocol from McCullum et al. [61] with our grafted nanobodies. | ||
| + | After testing the ideal annealing temperature for our oligonucleotides, we conducted the epPCR experiments and the amplification of the estradiol-nanobody-graft did work: | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild9.jpg|thumb|800px|center| Figure 9: Gel image of the by error prone PCR amplified estradiol nanobody fragment. The numbers 1 to 20 depict the dilution round the sample stems from. A 100bp plus ladder was used for size comparison. The gel (1.2% agarose, 1x TAE) was run for 25 minutes at 100 V. The amplified estradiol nanobody fragment is 400 bp large. The samples were run on two gels under the same conditions.]] | ||
| + | |||
| + | |||
| + | |||
| + | However, we had serious problems to amplify the sequence of the progesterone nanobody by epPCR: The epPCR did not work even after several runs. | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 10: Gel image of the by error prone PCR amplified progesterone nanobody fragment. The numbers 1 to 20 depict the dilution round the sample stems from. A 100bp plus ladder was used for size comparison. The gel (1.2% agarose, 1x TAE) was run for 25 minutes at 100 V. The amplified progesterone nanobody fragment is 400 bp large. The samples were run on two gels under the same conditions. A strong signal is only visible in the first two dilution steps. A weak signal can be seen until dilution 5, afterwards no bands can be seen anymore.]] | ||
| + | |||
| + | |||
| + | |||
| + | Therefore, we decided to purchase and use a commercial epPCR kit from JenaBioScience [16]. With this kit we were able to also amplify the sequence of the progesterone nanobody. | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild11.jpg|thumb|800px|center|Figure 11: Gel image of the by error prone PCR amplified nanobody fragments using the epPCR kit. 5 samples per nanobody were applied. A 100bp plus ladder was used for size comparison. The gel (1.2% agarose, 1x TAE) was run for 25 minutes at 100 V. The amplified nanobody fragments are ~400 bp large.]] | ||
| + | |||
| + | |||
| + | |||
| + | Sequencing | ||
| + | For our antibody-hormone binding process on the SAW chip we need a library of potential binders to find the best possible binder for our system. Therefore, we performed an error prone PCR (epPCR) of our estradiol and progesteron nanobody sequence to grant a high variability of binders. We chose to test two different epPCR systems to evaluate the best system for our approach. To determine the mutation frequency of the two epPCR systems, we decided to use Illumina MiSeq sequencing. For this examination we tested and compared four different samples: The estradiol nanobody amplificated by the epPCR protocol of McCullum et al. from 2010 [12] (sample “E2_S”), the estradiol nanobody (sample “E2_K”) and progesterone nanobody (sample “P_K”) amplified by epPCR using the kit from JenaBioScience [65] and as a reference the estradiol nanobody amplificated by a OneTaq® High Fidelity DNA polymerase. All four samples were sequenced by Illumina MiSeq technology to assess the diversity of the library as well as the mutation frequency. The library samples were first checked for fragment length and successful removal of the Illumina adapters. This was verified using the Agilent Bioanalyzer 2100 with the High Sensitivity DNA kit[17]. All samples showed an optimal library size and a complete removal of the unbound adapter fragments and were ready for the sequencing. Subsequently, the samples were sequenced. We chose a paired end approach with 250 bp per read to get a complete and high coverage of the 410 bp long nanobody sequence. | ||
| + | After the successful sequencing, the quality of the reads was checked with the tool FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and processed accordingly. Shown are the results of the sample E2_S. | ||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 12: Quality distribution of the reads from E2_S pre trimming. Analyzed with FastQC. Quality is measured with Phred score. Red area depicts a quality range from 0 to 20, yellow area from 21 to 28 and green area is a quality range from 29 to 38. The reads are represented by box plots: The average quality of the reads at each position is shown in the blue line. The median quality is represented by the red line. The yellow boxes indicate the quantiles and the extreme values are displayed by the black lines. It can be seen that the quality of the reads decreases with increasing length.]] | ||
| + | |||
| + | The first step of read processing is to filter the raw data for high quality reads and drop the reads with low quality. Therefore, we trimmed the reads with the tool Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic) by cutting off the first and last three bases as well as after four consecutive bases with a quality score lower than 20. If the reads still remained a size of 36 or more, they were kept. In this process 94-96% of the reads survived the trimming process. The trimmed library was analyzed as well with FastQC: | ||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 13: Quality distribution of the reads from E2_S post trimming. Analyzed with FastQC. Quality is measured with Phred score. The red area depicts a quality range from 0 to 20, yellow area from 21 to 28 and green area is a quality range from 29 to 38. The reads are represented by box plots: The average quality of the reads at each position is shown in the blue line. The median quality is represented by the red line. The yellow boxes indicate the quantiles and the extreme values are displayed by the black lines. It is evident that the quality improved significantly through processing.]] | ||
| + | |||
| + | As one can see, it is evident that the quality improved significantly through processing (see Fig. 11 and 12). After trimming, the length of reads, which are in terms of their quality, still in the green area increased from around 200 to 260 bp. Furthermore, the amount of reads in the red quality area decreased drastically and no quantile of reads reaches this area. | ||
| + | |||
| + | After trimming of the raw data, the reads were mapped on the associated reference genome (estradiol nanobody or progesterone nanobody). The Mapping was done with the mapping aligner Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). The important mapping statistics are collected with the command flagstat of the tool samtools and are shown in the following table: | ||
| + | Table 3: Flagstats for the four sequenced samples E2_O, E2_S, E2_K and P_K. The data was retrieved using the “flagstat” command from samtools analysing the with Bowtie2 mapped data. | ||
| + | Sample | ||
| + | attribute E2_O E2_S E2_K P_K | ||
| + | Reads in total 733474 882572 886832 948928 | ||
| + | Read 1 364161 437139 418312 458932 | ||
| + | Read 2 363718 436823 416450 457526 | ||
| + | Properly paired 726754 (99,85%) 872830 (99,87%) 829972 (99,43%) 912058 (99,52%) | ||
| + | Mapped reads 727879 (99,24%) 873962 (99,02%) 834762 (94,1%) 916458 (96,58%) | ||
| + | Nearly all reads were properly paired. | ||
| + | To visualize our data and check for the coverage of our samples, we chose the Integrative Genomic Viewer [18]. IGV is a tool to visualize genomic data and was used by us to compare the reference sequence (our initially grafted sequences) with the sequenced samples to see for mutations and visualize these. | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 14: Screenshot from the IGV file of the estradiol samples´ bam files, showing the coverage of the samples E2_O, E2_S and E2_K. Furthermore, deviations from the reference sequence (the initially grafted sequence of the estradiol nanobody) are shown and each base has a distinct colour: Adenine is shown in green, cytosine in blue, guanine in orange and thymine in red. The first paragraph depicts sample E2_O, second paragraph E2_S and the third paragraph E2_K.]] | ||
| + | |||
| + | As shown in Figure 14, the two epPCR approaches show significantly more mutations than the control. Furthermore, an even distribution of the mutation sites is shown. The most noticeable feature of the IGV visualization are the two cytosines between base 200 and 250, which are present throughout all samples. These two mutations are also present in the control sample E2_O, where otherwise significantly fewer mutations occurred. This indicates that an error occurred during gene synthesis. | ||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 15: Screenshot from the IGV file of the progesterone sample bam file, showing the coverage of the samples P_K. Furthermore, deviations from the reference sequence (initial sequence of the progesterone graft) are shown and each base has a distinct colour: Adenine is shown in green, cytosine in blue, guanine in orange and thymine in red. The first paragraph depicts the sample P_K.]] | ||
| + | |||
| + | |||
| + | |||
| + | Figure 15 shows an even distribution of the mutation sites. The same two cytosines between base 200 and 250, which were present throughout all samples from the estradiol samples (E2_O, E2_S and E2_K), are present in the P_K sample as well. These facts strengthen the assumption that an error occurred during gene synthesis. | ||
| + | For the analysis of the mutation rate of each sample, we have to call the base for each read on each position on the reference. For this purpose, we wrote a python script, which used the samtools command “mpileup” to get the coverage informations for each position and afterwards count all A’s, T’s, G’s and C’s. Lastly, it writes the position on the reference with the reference base, the total coverage and all counted bases from the different reads in one single excel document. | ||
| + | |||
| + | Table 4: Excel table created by the used python script, which shows the coverage of all reads per position. It also shows the percentage of each of the four bases per position in relation to the total number of reads. Shown are the first 10 of 410 lines of the excel sheet of the sample Prog_K. | ||
| + | Position Reference Sequence A C G T coverage | ||
| + | 1 A 339539 1294 1015 602 339129 | ||
| + | 2 T 1385 134 78 343302 344756 | ||
| + | 3 T 5490 291 157 361494 366775 | ||
| + | 4 A 367264 316 271 1410 367969 | ||
| + | 5 T 525 141 77 372295 372631 | ||
| + | 6 A 382438 357 378 2953 385930 | ||
| + | 7 G 494 435 395276 829 397093 | ||
| + | 8 G 627 1991 397089 1907 401218 | ||
| + | 9 C 191 413368 161 799 415012 | ||
| + | 10 C 267 414865 61 594 415740 | ||
| + | As can be seen in Table 4, coverage and reference sequence match. Deviations are only marginal. | ||
| + | With the detailed coverage information, it was possible to calculate the mutation frequency per base pair of all four samples. Therefore, we wrote a python script which imports an excel file, counts coverage for all non-reference bases at every position as well as the total coverage, normalizes the counted mutations by the total coverage of the analyzed sample and calculates the mutation frequency. These mutation frequency values are shown in the following table. | ||
| + | |||
| + | Table 5: Mutation frequencies [bp-1] for the four sequenced samples. | ||
| + | Sample Mutation frequency [bp]-1>/sup> | ||
| + | E2_O 3,302 * 10-4 | ||
| + | E2_S 1,992 * 10-3 | ||
| + | E2_K 1,101 * 10-3 | ||
| + | E2_P 1,283 * 10-3 | ||
| + | Using the data from table 5, the following graph was created. | ||
| + | |||
| + | Figure 16: Graphical display of the calculated mutation frequencies [bp-1] of the “selfmade” and the kit epPCR amplification. Shown are the estradiol nanobody control sample, amplified by OneTaq DNA polymerase (E2_O, in grey); the estradiol sample generated by epPCR amplification using the protocol of McCullum et al. from 2010 (E2_S, in dark red) and the two samples of estradiol (E2_K, in light red) and progesterone (P_K, in green) amplified using the epPCR kit bought from JenaBioScience. | ||
| + | |||
| + | The calculated mutation frequency for the control (E2_O) is 3,3 ×10-4 mutations per base pair, which is slightly higher than the expected sequencing error of 1.6×10-4 [19]. Since the mutation frequencies are of the same order of magnitude and the control is only slightly higher, the increased error rate cannot be explained by sequencing errors but by the natural error rate of the used OneTaq DNA Polymerase (<140*10-6 bases) [13]. | ||
| + | The “selfmade” approach using the epPCR protocol from McCullum et al. from 2010 with the OneTaq® DNA Polymerase yielded the highest rate of mutations per base pair and is six times higher compared to the control not using epPCR conditions. This strongly shows the negative effect of the parameters chosen (higher Mg2+ concentrations, presence of MnCl2+, more time for elongation, imbalanced rate of dNTPs and higher concentrastions of DNA polymerase) on the error rate of the used DNA polymerase. | ||
| + | |||
| + | The mutation rate for the JenaBioScience epPCR kit is expected to be between 0,6 to 2% [16]. In our experiments the kit yielded mutation rates per base pair of 1,101 * 10-3 for the estradiol nanobody and 1,283 * 10-3 for the progesterone nanobody, resulting in an average mutation rate per base pair of 1,192 * 10-3 per run of PCR. Although the “selfmade” epPCR protocol yielded a higher mutation rate per base pair we nevertheless chose the kit to generate our library from the nanobody fragments. This is because we could not achieve amplification of the progesterone nanobody by this protocol. That is why we chose the kit in the first place. | ||
| + | Cloning | ||
| + | After ensuring that random mutagenesis by epPCR had worked, we proceeded to clone the variable fragments into the phagemid vector pZMB0062. A restriction digest with SfiI [20] was used for cloning. SfiI produces two different sticky ends (see Figures 4 and 5), due to the non-palindromic recognition sequence (GGCCNNNNNGGCC), resulting in different sequences of the 5'- and 3'-cleavage sites. This also prevents a religation of the phagemid vector and enables an easy cloning of the variable sequences. | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 17: Overview of SfiI restriction sites. The gene of interest will be inserted between the pelB signal peptide (light pink coloured) and the linker (dark purple coloured), connecting it to the M13 geneIII protein. As insert is the grafted estradiol nanobody (grey coloured) and the immobilisation tag (green coloured) shown. A) Sequence of the 5´ SfiI restriction site with indicated cutting pattern. B) Sequence of the 3´ SfiI restriction site with indicated cutting pattern.]] | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 18: Map of the phagemid vector pZMB0062 with the sequence of the nanobody progesterone inserted.]] | ||
| + | |||
| + | Following the ligation, we transformed the E. coli strain ER2738 using the electroshock transformation protocol. All resulting clones were screened by colony PCR to verify the integration of the insert into the vector. | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 19: Colony PCR of ER2738 clones after one day of incubation at 37 °C. Colonies 2, 3, 4, 7, 8, 9, 11 and 13 are positive for the progesterone nanobody insert. Colonies 1, 5, 10, 14 and 15 carry the religated vector without an insert. The gel (1,2% agarose with 1x TAE) was run at 100 V for 20 minutes. A 100 bp ladder was used for size reference.]] | ||
| + | |||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 20: Colony PCR of ER2738 clones after one day of incubation at 37 °C. The colony tested was positive for the estradiol nanobody insert. The gel (1,2% agarose with 1x TAE) was run at 100 V for 20 minutes. A 100 bp ladder was used for size reference.]] | ||
| + | |||
| + | Phage ELISA | ||
| + | Due to the overall difficult situation with COVID-19, we only were able to test our original nanobody grafts on their binding affinity. This examination was performed with a Phage ELISA (enzyme linked immunosorbent assay). | ||
| + | |||
| + | For the detailed phage ELISA protocol click here. ---LINK ANLEGEN--- | ||
| + | |||
| + | A MaxiSorp 96-well plate was coated with streptavidin, subsequently biotin-coupled hormone was added to immobilize and present estradiol or progesterone. Incubation with our nanobodies presenting phages was followed by addition of anti-pVIII antibody with coupled HRP (horseradish peroxidase) and further addition of the substrate ABTS. Finally, the absorbance was measured in a Tecan i-control infinite 200 at 419 nm. | ||
| + | |||
| + | Table 6: Raw data (top) and corrected data (bottom) of the phage ELISA. The estradiol nanobody was tested against biotin conjugated estradiol in wells B4 to B6 and C4 to C6, well B7 served as control. The progesterone nanobody was tested in wells D4 to D6 and E4 to E6 against biotin conjugated progesterone, well C7 served as a control. The detection reaction was performed with the HRP conjugated anti pVIII antibody and ABTS substrate. Absorbance was measured at 419 nm in the plate reader Tecan i-control infinite 200. | ||
| + | Raw data 4 5 6 7 | ||
| + | B 0,0523 0,0533 0,0521 0,0521 | ||
| + | C 0,0525 0,0522 0,0512 0,051 | ||
| + | D 0,049 0,0515 0,0518 | ||
| + | E 0,0518 0,0524 0,0429 | ||
| + | Corrected data 4 5 6 7 | ||
| + | B 0,0002 0,0012 0 0 | ||
| + | C 0,0004 0,0001 -0,0009 0 | ||
| + | D -0,002 0,0005 0,0008 | ||
| + | E 0,0008 0,0014 -0,0081 | ||
| + | |||
| + | |||
| + | The data (see Table 6) show no significant absorption at 419 nm, neither for the estradiol nanobody nor for the progesterone nanobody. Thus, it can be concluded that, as assumed and predicted, the initial grafts of the two antibodies do not bind their respective antigens or bind them poorly and that their specificity needs to improve by appropriate affinity maturation measures. | ||
| + | |||
| + | Outlook | ||
| + | As the initial grafts are only poor binders, their affinity needs to be improved. Therefore, the phagemid display will be conducted: In a next first step, the cloning and transformation of our created epPCR library of the two nanobodies will be performed. To achieve all possible variants of the nanobody sequences, we will have to pick several colonies and perform the phagemid display. For the application, we will adapt the panning conditions step by step to the conditions prevailing in urine or salvia (pH, other components, etc.) after the first 3 panning rounds, since the nanobodies should ultimately function in urine and detect the hormones in urine samples too. In total, we plan to perform 7 panning rounds to obtain a good and specific affinity of the nanobody to estradiol and progesterone. The remaining candidates in the last round of the phagemid display will be sequenced and amplified. For purification of the nanobodiies, the IMPACT kit will be used: Using Through Gibson assembly, the nanobody sequence of our affinity maturated nanobodies is cloned into the expression vector pTXB1. The vector carries an intein tag, as well as a chitin-binding domain (CBD), which will be used for purification of the nanobody and the off cleavage of the tag from the protein [70]. The resulting nanobodies are going to be immobilized on the gold surface of the chip using their immobilization tag and experiments on durability and sensitivity will be performed. | ||
| + | scFv Grafting | ||
| + | Due to their small size, scFvs can easily be expressed as unique units or as fusion proteins in bacteria and also be immobilized on a surface with a high density [71]. To obtain our scFvs for binding the hormones progesterone and estradiol, we have linked the sequences of the light and heavy chain of estradiol [72] and progesterone antibodies [24] with a histidine linker with a length of 15 amino acids. The histidine linker between the chains was later used to immobilize the scFv on the SAW chip. | ||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 21: Visualization of the scFv Fragment for progesterone binding. The figure shows the overlap for Gibson Assembly at the 3’ and 5’ end of the fragment. Also the restrictionsites of SpeI and NdeI are shown.]] | ||
| + | |||
| + | For cloning the scFv fragments into the vector pTXB1 via Gibson Assembly®, 30 bp overhangs were added to the 3' and 5' end of the fragment. The cloning of the scFvs was initially tested successfully in silico. | ||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--Bild1.jpg|thumb|800px|center|Figure 22: : Map of the pTXB1 vector with inserted scFv progesterone fragment.]] | ||
| + | |||
| + | pTXB1 is an IPTG inducible expression vector with a size of ~ 6700 bp. It mediates an ampicillin resistance and is suitable for the in-frame insertion of a target gene upstream of a mxe intein protein. We choose this vector over using one of the iGEM standard backbones, due to the possibility to express tag-free. | ||
| + | |||
| + | The vector was used to overexpress our scFvs after successful cloning. Subsequently, the proteins can be isolated with the IMPACT™ Protein Purification System [70]. | ||
| + | scFv Cloning Strategies | ||
| + | For cloning our scFvs for binding of progsterone into the pTXB1 backbone, three different cloning strategies were applied and tested. The scFv fragment for progesterone binding was ordered as gene synthesis from IDT and Eurofins, already with overhangs complementary to the backbone. | ||
| + | Cloning via Gibson Assembly® | ||
| + | To clone the scFv fragment into the pTXB1 backbone via Gibson Assembly®, the pTXB1 backbone was opened by a PCR upstream of the Mxe Intein and thus linearized. Phusion DNA polymerase was used for the PCR, as it has a proof-reading function. The primers for opening the backbone mediated a 30 bp overhang complementary to the overhang of the scFv fragment. | ||
| + | |||
| + | 1.1 After linearization of the backbone, a gel electrophoresis was performed in which the complete PCR product was inserted. The gel was then stained in ethidium bromide. The resulting fragments were visible on the transiluminator afterwards. With a scalpel the band was cut out of the gel at the ~ 6700 bp mark. The DNA was then recovered by using the Gel Clean up Kit from Machery-Nagel. The isolated DNA fragment and the scFv were then used for the Gibson Assemly®. | ||
| + | |||
| + | 1.2 A second approach to preserve the opened backbone was the digestion of the PCR product by the restriction enzyme DpnI. Digestion with this restriction enzyme allows methylated template to be removed from the PCR, leaving only synthesized linearized plasmid DNA. After DpnI digestion, a PCR clean up was performed with the Machery-Nagel PCR and DNA clean up kit to remove debrist. The Gibson Assemly® followed. | ||
| + | Cloning via Seamless Cloning | ||
| + | In addition to the Gibson Assemly®, the Seamless Cloning method was tested. The Seamless Cloning Kit from Thermo Fisher [25] was used. The Invitrogen™ GeneArt™ Seamless Cloning and Assembly Kits allow in vitro cloning in nearly any linearized vector, in around 30 minutes, without additional DNA sequences, restriction endonucleases or ligation. In this method, the backbone was opened as already described for the Gibson assembly. The opened backbone and the scFv fragment are combined. Then, as with the Gibson Assembly®, the enzyme mix is added to the fragments to be cloned. The Invitrogen™ GeneArt™ Seamless Cloning and Assembly Kits allow in vitro cloning of up to 4 DNA fragments simultaneously in virtually any linearized vector, typically in 30 minutes, without additional DNA sequences, restrictions endonucleases or ligation. In this method, the backbone was opened as already described for the Gibson assembly. The opened backbone and the scFv fragment are combined. Then, as with the Gibson Assembly®, the enzyme mix is added to the fragments to be cloned together. | ||
| + | Cloning via Restriction Enzymes | ||
| + | The third cloning approach was a cloning using the restriction enzymes NdeI and SpeI, followed by a ligation with the T4 ligase. The backbone pTXB1 as well as the scFv fragment has a restriction site for both restriction enzymes. After the restriction the PCR mixture was purified with the PCR Clean Up Kit from Monarch. A ligation with the T4 ligase followed. | ||
| + | All cloning approaches were then transformed into E. coli DH5α. The resulting colonies were tested for the fragment by colony PCR. In case of a positive colony, plasmid isolation was performed with the Plasmidprep Kit from Thermo Fischer. The isolated plasmid was then delivered to the sequencing core facility of the CeBiTec for sequencing. | ||
| + | Outlook | ||
| + | After successful cloning of our scFv fragment into the expression vector pTXB1 with the goal to specifically bind progesterone, our next goal will be its expression, purification and subsequently immobilization of the ScFv protein on the SAW-chip surface. For the expression of the target fusion protein (scFv-Mxe), the plasmid pTXB1-scFv will be transformed into E. coli ER2566 and the expression is induced by Isopropyl-β-D-thiogalactopyranosid (IPTG). Due to the toxicity of IPTG, the cells are induced while the exponential phase of growth at the OD600 of 0.5. The expression of the protein may require the optimization of the expression conditions. Therefore, different cultivation temperatures can be compared and analyzed on their protein expression and its activity. We would have tested the protein expression at three different temperatures: 20°C, 30°C and 37°C. For further expression analysis, sampling will be performed after 3, 5, 8 and 16 hours of cultivation. After cultivation, the proteins will be purified using the IMPACT kit (NEB) [21]. This method is based on the principle of affinity chromaography. In order to verify the expression of the ScFv and its purification, protein concentration will be determined by Bradford assay and the protein expression and purification success of the different cultivation samples will be analyzed by SDS PAGE. Thereafter, the purified ScFv protein will be immobilized on the gold surface of the SAW-chip. For this purpose, we would have connected the isolated protein with the histidine in the linker to the gold surface [26]. After verification of the ScFv immobilization, the binding of our synthesized scFv to an horseradish peroxidase (HRP)-conjugated progesterone for improved photometrically detectability. Thereby, HRP oxidizes various organic and chromogenic substrates in the presence of hydrogen peroxide enabling the spectrophotometrically detection of the protein binding [67]. Thus, the binding ability of ScFv to progesterone is analyzed. The specific progesterone binding ScFv will be used for the SAW-chip based detection. | ||
| + | |||
References <br> | References <br> | ||
| Line 133: | Line 320: | ||
[49] S. R. Whaley, D. S. English, E. L. Hu, P. F. Barbara, und A. M. Belcher, „Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly“, Nature, Bd. 405, Nr. 6787, S. 665–668, Juni 2000, doi: 10.1038/35015043 Titel anhand dieser DOI in Citavi-Projekt übernehmen. | [49] S. R. Whaley, D. S. English, E. L. Hu, P. F. Barbara, und A. M. Belcher, „Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly“, Nature, Bd. 405, Nr. 6787, S. 665–668, Juni 2000, doi: 10.1038/35015043 Titel anhand dieser DOI in Citavi-Projekt übernehmen. | ||
| + | |||
| + | [50] M. Dondelinger u. a., „Understanding the Significance and Implications of Antibody Numbering and Antigen-Binding Surface/Residue Definition“, Front. Immunol., Bd. 9, 2018, doi: 10.3389/fimmu.2018.02278 Titel anhand dieser DOI in Citavi-Projekt übernehmen. | ||
| + | |||
| + | [51] A. Honegger und A. Plückthun, „Yet Another Numbering Scheme for Immunoglobulin Variable Domains: An Automatic Modeling and Analysis Tool“, J. Mol. Biol., Bd. 309, Nr. 3, S. 657–670, Juni 2001, doi: 10.1006/jmbi.2001.4662 Titel anhand dieser DOI in Citavi-Projekt übernehmen. | ||
| + | |||
| + | [52] R. C. Cadwell und G. F. Joyce, „Randomization of genes by PCR mutagenesis“, PCR Methods Appl., Bd. 2, Nr. 1, S. 28–33, Aug. 1992, doi: 10.1101/gr.2.1.28 Titel anhand dieser DOI in Citavi-Projekt übernehmen. | ||
| + | |||
| + | [53] E. O. McCullum, B. A. R. Williams, J. Zhang, und J. C. Chaput, „Random mutagenesis by error-prone PCR“, Methods Mol. Biol. Clifton NJ, Bd. 634, S. 103–109, 2010, doi: 10.1007/978-1-60761-652-8_7 Titel anhand dieser DOI in Citavi-Projekt übernehmen. | ||
| + | |||
| + | [54] N. E. Biolabs, „OneTaq® DNA Polymerase | NEB“. https://international.neb.com/products/m0480-onetaq-dna-polymerase (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [55] N. E. Biolabs, „Hot Start Taq DNA Polymerase | NEB“. https://international.neb.com/products/m0495-hot-start-taq-dna-polymerase (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [56] N. E. Biolabs, „Q5® High-Fidelity DNA Polymerase | NEB“. https://international.neb.com/products/m0491-q5-high-fidelity-dna-polymerase (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [57] J. B. GmbH, „JBS Error-Prone Kit, Molecular Evolution: Random Mutagenesis“, Jena Bioscience. https://www.jenabioscience.com/molecular-biology/cloning-and-mutagenesis/random-mutagenesis-kits/pp-102-jbs-error-prone-kit (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [58] „Quality control NGS libraries, Bioanalyzer High Sensitivity DNA | Agilent“. https://www.agilent.com/en/product/automated-electrophoresis/bioanalyzer-systems/bioanalyzer-dna-kits-reagents/bioanalyzer-high-sensitivity-dna-analysis-228249#productdetails (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [59] „igvtools | Integrative Genomics Viewer“. https://software.broadinstitute.org/software/igv/igvtools (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [60] „Understanding Illumina Quality Scores“, S. 2. | ||
| + | |||
| + | [61] N. E. Biolabs, „SfiI | NEB“. https://international.neb.com/products/r0123-sfii (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [62] N. E. Biolabs, „Impact | NEB“. https://international.neb.com/products/protein-expression-and-purification-technologies/e-coli/impact/impact (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [63] „Engineered Recombinant Single-Chain Fragment Variable Antibody for Immunosensors | Analytical Chemistry“. https://pubs.acs.org/doi/pdf/10.1021/ac0507690 Titel anhand dieser DOI in Citavi-Projekt übernehmen (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [64] R. P. D. Bank, „1D PFV: 1JNH“. https://www.rcsb.org/sequence/1JNH (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [65] R. P. D. Bank, „1D PFV: 1DBB“. https://www.rcsb.org/sequence/1DBB (zugegriffen Okt. 28, 2020). | ||
| + | |||
| + | [66] „geneart-seamless-cloning-assembly-mutagenesis-tools-brochure.pdf“. Zugegriffen: Okt. 28, 2020. [Online]. Verfügbar unter: https://tools.thermofisher.com/content/sfs/brochures/geneart-seamless-cloning-assembly-mutagenesis-tools-brochure.pdf. | ||
| + | |||
| + | [67] Z. Shen, G. A. Stryker, R. L. Mernaugh, L. Yu, H. Yan, und X. Zeng, „Single-Chain Fragment Variable Antibody Piezoimmunosensors“, Anal. Chem., Bd. 77, Nr. 3, S. 797–805, Feb. 2005, doi: 10.1021/ac048655w Titel anhand dieser DOI in Citavi-Projekt übernehmen. | ||
| + | |||
| + | [68] A. M. Azevedo, V. C. Martins, D. M. Prazeres, V. Vojinović, J. M. Cabral, und L. P. Fonseca, „Horseradish peroxidase: a valuable tool in biotechnology“, Biotechnol. Annu. Rev., Bd. 9, S. 199–247, 2003, doi: 10.1016/s1387-2656(03)09003-3 Titel anhand dieser DOI in Citavi-Projekt übernehmen. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 03:23, 28 October 2020
Nanobody scaffold suitable for grafting.
cAbBCII-10 is a nanobody, originally isolated as an antibody capturing and neutralizing enzymes of the β-lactamase B class [1].
As CDR acceptor (also called "scaffold") we used the nanobody cAbBCII-10 [1] (or also NbBcII10 [2] or 3DWT [3], from here on called "3DWT"). This Nanobody is also available as part of a composite part in the iGEM parts reg (BBa_K2082001). However, the nanobody is not available as basic part, so we also contributed in submitting the basic part of cAbBCII-10 (BBa_K3410001). We used the sequence deposited in the PDB by Vincke et al. [2, 3].
Originally, 3DWT was isolated as an antibody capturing and neutralizing enzymes of the β-lactamase B class [1].
A single-domain antibody, also known as nanobody, is an antibody consisting of a single variable antibody domain. First described in 1993 [6], nanobodies are an important research field for multiple pharmaceutical applications today. Mainly, nanobodies are divided in two types of different structures. The most common nanobodies are the VHH fragments, which consist only of a variable domain of a heavy chain and are mostly found in Camelidae species [8]. Another type of nanobodies are the VNAR fragments based on the IgNAR (“Immunoglobin new antigen receptor”) from cartilaginous fish [9]. Having a molecular mass of ∼12 kDa, the vNAR domain is the smallest antibody-like antigen binding domain known so far in the animals [10]. That is why VNAR domains have only two complementarity determining regions CDR1 and CDR3, in contrast to variable mammalian domains [11].
Accordingly, nanobodies are the smallest known antigen-binding proteins with a length of 4 nm and a diameter of 2.5 nm [12]. However, like a whole antibody, they are still able to recognize and bind specifically their target molecules using the single variable domain. Their compact structure and light molecular weight of only 12-15 kDa (compared to conventional antibodies with 150-160 kDa) make nanobodies the smallest active antigen-binding fragments [13]. Due to their versatile binding properties, high stability, manipulable characteristics as well as improved tissue penetration ability the nanobodies are crucial in the field of biotechnology or medicine today [14], [15].
Antibodies consist of two glycosylated heavy chains (H) with a molecular weight of 50 kDA each and two light chains of 25 kDA each, which are covalently linked via disulfide bridges. [16]. On the one hand, two N-terminal variable domains (V) of the heavy (VH) and light chain (VL) form the antigen-binding domain, also called paratrope. On the other hand, the C-terminal regions form the three constant domains CH1, CH2 and CH3. Compared to common antibodies, the nanobody family consist of conventional heterotetramic antibodies with low-affinity binders and as well as unique functional heavy(H)-chain antibodies (HCAbs) with high affinity binders [13]. The H chain of these homodimeric antibodies consist of one antigen-binding domain, the VHH, and two constant domains. The HCAbs fail to incorporate light(L)-chains owing to the deletion of the first constant domain and a reshaped surface of the VHH side, which normally associates with the L-chain in common antibodies [17]. This structural change leads to this enlargement allowing the nanobodies to bind their antigen with a high affinity Furthermore, the hydrophobic amino acids within the conserved region (frame region; FR), which ensure the interaction of VH and VL, are replaced by hydrophilic amino acids [12], [18].
The field of application of nanobodies is very diverse. Their use in diagnostic test systems and therapeutic options is increasing. In contrast to conventional antibodies, nanobodies merge as a new technical achievement due to their low molecular weight (half the size of single-chain variable elements and ten times lighter than conventional antibodies) and small size (2-3 nm) [17]. Moreover, a high expression rate of nanobodies was shown in E. coli with high functionality and stability even in absence of conserved disulphide bonds [19]. The loss of the L-chain leads to missing disulphide modifications in nanobodies, which makes them suitable for production in different bacteria and yeast [20], [21]. Additionally, their high tolerance against acid conditions and temperature compared to mouse monoclonal antibodies [8], high solubility due to a tetrahedron of highly conserved hydrophilic substitutions [22] and very few cleavage sites for enzymes [13] make the nanobodies very promising and an interesting research subject for new applications in the biotechnology. Furthermore, the recombinant nature and single domain of the nanobodies allows easy generation, production and molecular biological manipulation. These include sequential modifications, the generation of fusion proteins with other substances (e.g. pharmaceuticals), the transfer of antigen specificities as well as the transfer of affinity from one nanobody to another [23].
Due to their low mass, the nanobodies are able to easily cross the blood-brain barrier and make them able to quickly leave the human body's circulation via urine after resolving their function inside the cell. Furthermore, nanobodies show a higher availability to tissue penetration and due to their smaller size, they trigger less immunological reaction, which leads to better pharmacokinetics [24]. All stated points are an important property for therapeutic options (cancer therapies) and diagnostic processes (molecular imaging) [23]. Nanobody Grafting Typically, a camelid immune library is needed for the generation of nanobodies, besides ethical concerns, you would have to keep animals and take care of them. One can also obtain nanobodies from synthetic libraries, but this requires large libraries and sophisticated selection mechanisms. An alternative is the so-called grafting of nanobodies. In this method, the CDRs of any antibody are transferred to an existing nanobody to create a mixed nanobody [4]. However, not only the three CDR regions 1 to 3 are transferred, but also amino acids important for the structure and function are transferred from the donor antibody to the nanobody scaffold [25].

The grafting itself is carried out in silico, therefore the amino acid residues from the VH domain from the donor antibody and from the accepting nanobody have to be numbered according to a numbering scheme like Kabat or AHo [26] and CDR1 to 3 have to be defined accordingly [25]. The correct structure of the graft can be checked with the help of visualization programs such as ChimeraX [27]. Here you can see the results of our grafting. Error Prone PCR For means of affinity maturation we needed a library of our grafted sequence. To introduce the required diversity into the grafted sequence, we chose a random mutagenesis approach by polymerase chain reaction (PCR), first described by Cadwell and Joyce in 1992 [28]. This variant of the PCR-technique is termed error prone PCR (epPCR). In this technique, the natural error rate of the Taq polymerase is increased by unfavorable reaction conditions. This is accomplished by: 1) Increased concentration of Taq DNA polymerase; 2) extended polymerase extension time; 3) increased concentration of MgCl2 ions; 4) unbalanced rate of dNTPs; and 5) addition of MnCl2 ions [29]. But how does grafting of antibodies work? A quick guide how to graft:
In a first step of in silico nanobody grafting, the amino acid sequences of both donor (10G6D6 and C12G11, respectively) and acceptor (3DWT) are numbered. There are different numbering schemes for immunoglobulin variable domains available [9], we decided to use the AHo numbering scheme, because placement of gaps is based on the spatial alignment of known three dimensional structures of immunoglobulin domains and not on sequence variability [10]. The numbering was performed using ANARCI (SAbPred) (http://opig.stats.ox.ac.uk/webapps/newsabdab/sabpred/anarci/). For further work, the numbered residues were transferred to an excel sheet. Both, the CDRs and the functional important framework residues of the donor (10G6D6 and C12G11) were combined with the acceptor (3DWT) and a new nanobody was created (Graft).


The grafting results with the exact sequence can be found in table 1 for our estradiol nanobody and in table 2 for our progesterone nanobody. Table 1: Design of the estradiol nanobody. The first 44 amino acid residues (AHo numbering) of the donor antibody (10G6), the accepting antibody (3DWT) and the grafted estradiol nanobody are shown. The complementarity determining regions are highlighted in yellow.
[[File:T--Bielefeld-CeBiTec--Bild6.jpg|thumb|800px|center|Figure 6: Design of the progesterone nanobody. The first 44 amino acid residues (AHo numbering) of the donor antibody (C12G11), the accepting antibody (3DWT) and the grafted progesterone nanobody are shown. The complementarity determining regions are highlighted in yellow.
Using ChimeraX (https://www.cgl.ucsf.edu/chimerax/), we optically verified the 3D structure of the grafted nanobodies for steric obstacles to antigen binding: For example, we checked, if the grafted residues sterically impaired residues from the framework.
Subsequently, the amino acid sequences were translated into a complementary DNA sequence. This was performed using the tool Sequence Manipulation Suite (http://bioinformatics.org/sms2/rev_trans.html).
Furthermore, the sequences were optimized in regard of their GC content and number of repeats as well as their length. The created nucleotide sequences were purchased as gene synthesis from IDT and EurofinsGenomics. In general, the nanobody grafting process generates weak binders as the sequence of the scaffold is strongly modified, especially with regard to the CDRs, which can partly form new secondary structures. In addition, core frame work residues are also transferred from the donor to the scaffold, which changes its sequence, perhaps resulting in a changed structure of the scaffold in non-CDR areas. Therefore, the grafted nanobodies have to be improved by suitable affinity maturation measures [9].
Because the epPCR conducted with the OneTaq® DNA Polymerase showed better results regarding positive clones (gelbild colony PCR (see Fig. 6) as well as more and larger red colonies after transformation (see Fig. 3) compared to the results of the Taq polymerase, we decided to use OneTaq® DNA Polymerase for our epPCR experiments using the protocol from McCullum et al. [61] with our grafted nanobodies.
After testing the ideal annealing temperature for our oligonucleotides, we conducted the epPCR experiments and the amplification of the estradiol-nanobody-graft did work:

However, we had serious problems to amplify the sequence of the progesterone nanobody by epPCR: The epPCR did not work even after several runs.

Therefore, we decided to purchase and use a commercial epPCR kit from JenaBioScience [16]. With this kit we were able to also amplify the sequence of the progesterone nanobody.
Sequencing For our antibody-hormone binding process on the SAW chip we need a library of potential binders to find the best possible binder for our system. Therefore, we performed an error prone PCR (epPCR) of our estradiol and progesteron nanobody sequence to grant a high variability of binders. We chose to test two different epPCR systems to evaluate the best system for our approach. To determine the mutation frequency of the two epPCR systems, we decided to use Illumina MiSeq sequencing. For this examination we tested and compared four different samples: The estradiol nanobody amplificated by the epPCR protocol of McCullum et al. from 2010 [12] (sample “E2_S”), the estradiol nanobody (sample “E2_K”) and progesterone nanobody (sample “P_K”) amplified by epPCR using the kit from JenaBioScience [65] and as a reference the estradiol nanobody amplificated by a OneTaq® High Fidelity DNA polymerase. All four samples were sequenced by Illumina MiSeq technology to assess the diversity of the library as well as the mutation frequency. The library samples were first checked for fragment length and successful removal of the Illumina adapters. This was verified using the Agilent Bioanalyzer 2100 with the High Sensitivity DNA kit[17]. All samples showed an optimal library size and a complete removal of the unbound adapter fragments and were ready for the sequencing. Subsequently, the samples were sequenced. We chose a paired end approach with 250 bp per read to get a complete and high coverage of the 410 bp long nanobody sequence. After the successful sequencing, the quality of the reads was checked with the tool FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and processed accordingly. Shown are the results of the sample E2_S.

The first step of read processing is to filter the raw data for high quality reads and drop the reads with low quality. Therefore, we trimmed the reads with the tool Trimmomatic (http://www.usadellab.org/cms/?page=trimmomatic) by cutting off the first and last three bases as well as after four consecutive bases with a quality score lower than 20. If the reads still remained a size of 36 or more, they were kept. In this process 94-96% of the reads survived the trimming process. The trimmed library was analyzed as well with FastQC:

As one can see, it is evident that the quality improved significantly through processing (see Fig. 11 and 12). After trimming, the length of reads, which are in terms of their quality, still in the green area increased from around 200 to 260 bp. Furthermore, the amount of reads in the red quality area decreased drastically and no quantile of reads reaches this area.
After trimming of the raw data, the reads were mapped on the associated reference genome (estradiol nanobody or progesterone nanobody). The Mapping was done with the mapping aligner Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). The important mapping statistics are collected with the command flagstat of the tool samtools and are shown in the following table: Table 3: Flagstats for the four sequenced samples E2_O, E2_S, E2_K and P_K. The data was retrieved using the “flagstat” command from samtools analysing the with Bowtie2 mapped data. Sample attribute E2_O E2_S E2_K P_K Reads in total 733474 882572 886832 948928 Read 1 364161 437139 418312 458932 Read 2 363718 436823 416450 457526 Properly paired 726754 (99,85%) 872830 (99,87%) 829972 (99,43%) 912058 (99,52%) Mapped reads 727879 (99,24%) 873962 (99,02%) 834762 (94,1%) 916458 (96,58%) Nearly all reads were properly paired. To visualize our data and check for the coverage of our samples, we chose the Integrative Genomic Viewer [18]. IGV is a tool to visualize genomic data and was used by us to compare the reference sequence (our initially grafted sequences) with the sequenced samples to see for mutations and visualize these.

As shown in Figure 14, the two epPCR approaches show significantly more mutations than the control. Furthermore, an even distribution of the mutation sites is shown. The most noticeable feature of the IGV visualization are the two cytosines between base 200 and 250, which are present throughout all samples. These two mutations are also present in the control sample E2_O, where otherwise significantly fewer mutations occurred. This indicates that an error occurred during gene synthesis.

Figure 15 shows an even distribution of the mutation sites. The same two cytosines between base 200 and 250, which were present throughout all samples from the estradiol samples (E2_O, E2_S and E2_K), are present in the P_K sample as well. These facts strengthen the assumption that an error occurred during gene synthesis. For the analysis of the mutation rate of each sample, we have to call the base for each read on each position on the reference. For this purpose, we wrote a python script, which used the samtools command “mpileup” to get the coverage informations for each position and afterwards count all A’s, T’s, G’s and C’s. Lastly, it writes the position on the reference with the reference base, the total coverage and all counted bases from the different reads in one single excel document.
Table 4: Excel table created by the used python script, which shows the coverage of all reads per position. It also shows the percentage of each of the four bases per position in relation to the total number of reads. Shown are the first 10 of 410 lines of the excel sheet of the sample Prog_K. Position Reference Sequence A C G T coverage 1 A 339539 1294 1015 602 339129 2 T 1385 134 78 343302 344756 3 T 5490 291 157 361494 366775 4 A 367264 316 271 1410 367969 5 T 525 141 77 372295 372631 6 A 382438 357 378 2953 385930 7 G 494 435 395276 829 397093 8 G 627 1991 397089 1907 401218 9 C 191 413368 161 799 415012 10 C 267 414865 61 594 415740 As can be seen in Table 4, coverage and reference sequence match. Deviations are only marginal. With the detailed coverage information, it was possible to calculate the mutation frequency per base pair of all four samples. Therefore, we wrote a python script which imports an excel file, counts coverage for all non-reference bases at every position as well as the total coverage, normalizes the counted mutations by the total coverage of the analyzed sample and calculates the mutation frequency. These mutation frequency values are shown in the following table.
Table 5: Mutation frequencies [bp-1] for the four sequenced samples. Sample Mutation frequency [bp]-1>/sup> E2_O 3,302 * 10-4 E2_S 1,992 * 10-3 E2_K 1,101 * 10-3 E2_P 1,283 * 10-3 Using the data from table 5, the following graph was created.
Figure 16: Graphical display of the calculated mutation frequencies [bp-1] of the “selfmade” and the kit epPCR amplification. Shown are the estradiol nanobody control sample, amplified by OneTaq DNA polymerase (E2_O, in grey); the estradiol sample generated by epPCR amplification using the protocol of McCullum et al. from 2010 (E2_S, in dark red) and the two samples of estradiol (E2_K, in light red) and progesterone (P_K, in green) amplified using the epPCR kit bought from JenaBioScience.
The calculated mutation frequency for the control (E2_O) is 3,3 ×10-4 mutations per base pair, which is slightly higher than the expected sequencing error of 1.6×10-4 [19]. Since the mutation frequencies are of the same order of magnitude and the control is only slightly higher, the increased error rate cannot be explained by sequencing errors but by the natural error rate of the used OneTaq DNA Polymerase (<140*10-6 bases) [13]. The “selfmade” approach using the epPCR protocol from McCullum et al. from 2010 with the OneTaq® DNA Polymerase yielded the highest rate of mutations per base pair and is six times higher compared to the control not using epPCR conditions. This strongly shows the negative effect of the parameters chosen (higher Mg2+ concentrations, presence of MnCl2+, more time for elongation, imbalanced rate of dNTPs and higher concentrastions of DNA polymerase) on the error rate of the used DNA polymerase.
The mutation rate for the JenaBioScience epPCR kit is expected to be between 0,6 to 2% [16]. In our experiments the kit yielded mutation rates per base pair of 1,101 * 10-3 for the estradiol nanobody and 1,283 * 10-3 for the progesterone nanobody, resulting in an average mutation rate per base pair of 1,192 * 10-3 per run of PCR. Although the “selfmade” epPCR protocol yielded a higher mutation rate per base pair we nevertheless chose the kit to generate our library from the nanobody fragments. This is because we could not achieve amplification of the progesterone nanobody by this protocol. That is why we chose the kit in the first place. Cloning After ensuring that random mutagenesis by epPCR had worked, we proceeded to clone the variable fragments into the phagemid vector pZMB0062. A restriction digest with SfiI [20] was used for cloning. SfiI produces two different sticky ends (see Figures 4 and 5), due to the non-palindromic recognition sequence (GGCCNNNNNGGCC), resulting in different sequences of the 5'- and 3'-cleavage sites. This also prevents a religation of the phagemid vector and enables an easy cloning of the variable sequences.

Following the ligation, we transformed the E. coli strain ER2738 using the electroshock transformation protocol. All resulting clones were screened by colony PCR to verify the integration of the insert into the vector.

Phage ELISA Due to the overall difficult situation with COVID-19, we only were able to test our original nanobody grafts on their binding affinity. This examination was performed with a Phage ELISA (enzyme linked immunosorbent assay).
For the detailed phage ELISA protocol click here. ---LINK ANLEGEN---
A MaxiSorp 96-well plate was coated with streptavidin, subsequently biotin-coupled hormone was added to immobilize and present estradiol or progesterone. Incubation with our nanobodies presenting phages was followed by addition of anti-pVIII antibody with coupled HRP (horseradish peroxidase) and further addition of the substrate ABTS. Finally, the absorbance was measured in a Tecan i-control infinite 200 at 419 nm.
Table 6: Raw data (top) and corrected data (bottom) of the phage ELISA. The estradiol nanobody was tested against biotin conjugated estradiol in wells B4 to B6 and C4 to C6, well B7 served as control. The progesterone nanobody was tested in wells D4 to D6 and E4 to E6 against biotin conjugated progesterone, well C7 served as a control. The detection reaction was performed with the HRP conjugated anti pVIII antibody and ABTS substrate. Absorbance was measured at 419 nm in the plate reader Tecan i-control infinite 200. Raw data 4 5 6 7 B 0,0523 0,0533 0,0521 0,0521 C 0,0525 0,0522 0,0512 0,051 D 0,049 0,0515 0,0518 E 0,0518 0,0524 0,0429 Corrected data 4 5 6 7 B 0,0002 0,0012 0 0 C 0,0004 0,0001 -0,0009 0 D -0,002 0,0005 0,0008 E 0,0008 0,0014 -0,0081
The data (see Table 6) show no significant absorption at 419 nm, neither for the estradiol nanobody nor for the progesterone nanobody. Thus, it can be concluded that, as assumed and predicted, the initial grafts of the two antibodies do not bind their respective antigens or bind them poorly and that their specificity needs to improve by appropriate affinity maturation measures.
Outlook As the initial grafts are only poor binders, their affinity needs to be improved. Therefore, the phagemid display will be conducted: In a next first step, the cloning and transformation of our created epPCR library of the two nanobodies will be performed. To achieve all possible variants of the nanobody sequences, we will have to pick several colonies and perform the phagemid display. For the application, we will adapt the panning conditions step by step to the conditions prevailing in urine or salvia (pH, other components, etc.) after the first 3 panning rounds, since the nanobodies should ultimately function in urine and detect the hormones in urine samples too. In total, we plan to perform 7 panning rounds to obtain a good and specific affinity of the nanobody to estradiol and progesterone. The remaining candidates in the last round of the phagemid display will be sequenced and amplified. For purification of the nanobodiies, the IMPACT kit will be used: Using Through Gibson assembly, the nanobody sequence of our affinity maturated nanobodies is cloned into the expression vector pTXB1. The vector carries an intein tag, as well as a chitin-binding domain (CBD), which will be used for purification of the nanobody and the off cleavage of the tag from the protein [70]. The resulting nanobodies are going to be immobilized on the gold surface of the chip using their immobilization tag and experiments on durability and sensitivity will be performed. scFv Grafting Due to their small size, scFvs can easily be expressed as unique units or as fusion proteins in bacteria and also be immobilized on a surface with a high density [71]. To obtain our scFvs for binding the hormones progesterone and estradiol, we have linked the sequences of the light and heavy chain of estradiol [72] and progesterone antibodies [24] with a histidine linker with a length of 15 amino acids. The histidine linker between the chains was later used to immobilize the scFv on the SAW chip.
For cloning the scFv fragments into the vector pTXB1 via Gibson Assembly®, 30 bp overhangs were added to the 3' and 5' end of the fragment. The cloning of the scFvs was initially tested successfully in silico.
pTXB1 is an IPTG inducible expression vector with a size of ~ 6700 bp. It mediates an ampicillin resistance and is suitable for the in-frame insertion of a target gene upstream of a mxe intein protein. We choose this vector over using one of the iGEM standard backbones, due to the possibility to express tag-free.
The vector was used to overexpress our scFvs after successful cloning. Subsequently, the proteins can be isolated with the IMPACT™ Protein Purification System [70]. scFv Cloning Strategies For cloning our scFvs for binding of progsterone into the pTXB1 backbone, three different cloning strategies were applied and tested. The scFv fragment for progesterone binding was ordered as gene synthesis from IDT and Eurofins, already with overhangs complementary to the backbone. Cloning via Gibson Assembly® To clone the scFv fragment into the pTXB1 backbone via Gibson Assembly®, the pTXB1 backbone was opened by a PCR upstream of the Mxe Intein and thus linearized. Phusion DNA polymerase was used for the PCR, as it has a proof-reading function. The primers for opening the backbone mediated a 30 bp overhang complementary to the overhang of the scFv fragment.
1.1 After linearization of the backbone, a gel electrophoresis was performed in which the complete PCR product was inserted. The gel was then stained in ethidium bromide. The resulting fragments were visible on the transiluminator afterwards. With a scalpel the band was cut out of the gel at the ~ 6700 bp mark. The DNA was then recovered by using the Gel Clean up Kit from Machery-Nagel. The isolated DNA fragment and the scFv were then used for the Gibson Assemly®.
1.2 A second approach to preserve the opened backbone was the digestion of the PCR product by the restriction enzyme DpnI. Digestion with this restriction enzyme allows methylated template to be removed from the PCR, leaving only synthesized linearized plasmid DNA. After DpnI digestion, a PCR clean up was performed with the Machery-Nagel PCR and DNA clean up kit to remove debrist. The Gibson Assemly® followed. Cloning via Seamless Cloning In addition to the Gibson Assemly®, the Seamless Cloning method was tested. The Seamless Cloning Kit from Thermo Fisher [25] was used. The Invitrogen™ GeneArt™ Seamless Cloning and Assembly Kits allow in vitro cloning in nearly any linearized vector, in around 30 minutes, without additional DNA sequences, restriction endonucleases or ligation. In this method, the backbone was opened as already described for the Gibson assembly. The opened backbone and the scFv fragment are combined. Then, as with the Gibson Assembly®, the enzyme mix is added to the fragments to be cloned. The Invitrogen™ GeneArt™ Seamless Cloning and Assembly Kits allow in vitro cloning of up to 4 DNA fragments simultaneously in virtually any linearized vector, typically in 30 minutes, without additional DNA sequences, restrictions endonucleases or ligation. In this method, the backbone was opened as already described for the Gibson assembly. The opened backbone and the scFv fragment are combined. Then, as with the Gibson Assembly®, the enzyme mix is added to the fragments to be cloned together. Cloning via Restriction Enzymes The third cloning approach was a cloning using the restriction enzymes NdeI and SpeI, followed by a ligation with the T4 ligase. The backbone pTXB1 as well as the scFv fragment has a restriction site for both restriction enzymes. After the restriction the PCR mixture was purified with the PCR Clean Up Kit from Monarch. A ligation with the T4 ligase followed. All cloning approaches were then transformed into E. coli DH5α. The resulting colonies were tested for the fragment by colony PCR. In case of a positive colony, plasmid isolation was performed with the Plasmidprep Kit from Thermo Fischer. The isolated plasmid was then delivered to the sequencing core facility of the CeBiTec for sequencing. Outlook After successful cloning of our scFv fragment into the expression vector pTXB1 with the goal to specifically bind progesterone, our next goal will be its expression, purification and subsequently immobilization of the ScFv protein on the SAW-chip surface. For the expression of the target fusion protein (scFv-Mxe), the plasmid pTXB1-scFv will be transformed into E. coli ER2566 and the expression is induced by Isopropyl-β-D-thiogalactopyranosid (IPTG). Due to the toxicity of IPTG, the cells are induced while the exponential phase of growth at the OD600 of 0.5. The expression of the protein may require the optimization of the expression conditions. Therefore, different cultivation temperatures can be compared and analyzed on their protein expression and its activity. We would have tested the protein expression at three different temperatures: 20°C, 30°C and 37°C. For further expression analysis, sampling will be performed after 3, 5, 8 and 16 hours of cultivation. After cultivation, the proteins will be purified using the IMPACT kit (NEB) [21]. This method is based on the principle of affinity chromaography. In order to verify the expression of the ScFv and its purification, protein concentration will be determined by Bradford assay and the protein expression and purification success of the different cultivation samples will be analyzed by SDS PAGE. Thereafter, the purified ScFv protein will be immobilized on the gold surface of the SAW-chip. For this purpose, we would have connected the isolated protein with the histidine in the linker to the gold surface [26]. After verification of the ScFv immobilization, the binding of our synthesized scFv to an horseradish peroxidase (HRP)-conjugated progesterone for improved photometrically detectability. Thereby, HRP oxidizes various organic and chromogenic substrates in the presence of hydrogen peroxide enabling the spectrophotometrically detection of the protein binding [67]. Thus, the binding ability of ScFv to progesterone is analyzed. The specific progesterone binding ScFv will be used for the SAW-chip based detection.
References
[1] K. E. Conrath et al., “Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae,” Antimicrobial Agents and Chemotherapy, vol. 45, no. 10, pp. 2807–2812, 2001, doi: 10.1128/AAC.45.10.2807-2812.2001.
[2] C. Vincke, R. Loris, D. Saerens, S. Martinez-Rodriguez, S. Muyldermans, and K. Conrath, “General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold,” The Journal of biological chemistry, vol. 284, no. 5, pp. 3273–3284, 2009, doi: 10.1074/jbc.M806889200.
[3] C. e. a. Vincke, 3DWT: Structure of CabBCII-10 nanobody.
[4] S. W. Fanning and J. R. Horn, “An anti-hapten camelid antibody reveals a cryptic binding site with significant energetic contributions from a nonhypervariable loop,” Protein science : a publication of the Protein Society, vol. 20, no. 7, pp. 1196–1207, 2011, doi: 10.1002/pro.648.
References
[1] K. E. Conrath et al., “Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae,” Antimicrobial Agents and Chemotherapy, vol. 45, no. 10, pp. 2807–2812, 2001, doi: 10.1128/AAC.45.10.2807-2812.2001.
[2] C. Vincke, R. Loris, D. Saerens, S. Martinez-Rodriguez, S. Muyldermans, and K. Conrath, “General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold,” The Journal of biological chemistry, vol. 284, no. 5, pp. 3273–3284, 2009, doi: 10.1074/jbc.M806889200.
[3] C. e. a. Vincke, 3DWT: Structure of CabBCII-10 nanobody.
[4] S. W. Fanning and J. R. Horn, “An anti-hapten camelid antibody reveals a cryptic binding site with significant energetic contributions from a nonhypervariable loop,” Protein science : a publication of the Protein Society, vol. 20, no. 7, pp. 1196–1207, 2011, doi: 10.1002/pro.648.
[5] D. Saerens et al., “Identification of a universal VHH framework to graft non-canonical antigen-binding loops of camel single-domain antibodies,” Journal of molecular biology, vol. 352, no. 3, pp. 597–607, 2005, doi: 10.1016/j.jmb.2005.07.038.
[6] C. Hamers-Casterman u. a., „Naturally occurring antibodies devoid of light chains“, Nature, Bd. 363, Nr. 6428, S. 446–448, Juni 1993, doi: 10.1038/363446a0 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[7] A. L. Nelson, „Antibody fragments“, mAbs, Bd. 2, Nr. 1, S. 77–83, 2010.
[8] R. H. van der Linden u. a., „Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies“, Biochim. Biophys. Acta, Bd. 1431, Nr. 1, S. 37–46, Apr. 1999, doi: 10.1016/s0167-4838(99)00030-8 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[9] R. L. Stanfield, H. Dooley, M. F. Flajnik, und I. A. Wilson, „Crystal structure of a shark single-domain antibody V region in complex with lysozyme“, Science, Bd. 305, Nr. 5691, S. 1770–1773, Sep. 2004, doi: 10.1126/science.1101148 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[10] C. Barelle, D. S. Gill, und K. Charlton, „Shark novel antigen receptors--the next generation of biologic therapeutics?“, Adv. Exp. Med. Biol., Bd. 655, S. 49–62, 2009, doi: 10.1007/978-1-4419-1132-2_6 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[11] S. Zielonka, M. Empting, J. Grzeschik, D. Könning, C. J. Barelle, und H. Kolmar, „Structural insights and biomedical potential of IgNAR scaffolds from sharks“, mAbs, Bd. 7, Nr. 1, S. 15–25, Dez. 2014, doi: 10.4161/19420862.2015.989032 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[12] W. Wang, J. Yuan, und C. Jiang, „Applications of nanobodies in plant science and biotechnology“, Plant Mol. Biol., Okt. 2020, doi: 10.1007/s11103-020-01082-z Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[13] M. M. Harmsen und H. J. De Haard, „Properties, production, and applications of camelid single-domain antibody fragments“, Appl. Microbiol. Biotechnol., Bd. 77, Nr. 1, S. 13–22, Nov. 2007, doi: 10.1007/s00253-007-1142-2 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[14] P. Chames und U. Rothbauer, „Special Issue: Nanobody“, Antibodies, Bd. 9, Nr. 1, Art. Nr. 1, März 2020, doi: 10.3390/antib9010006 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[15] X. Yu u. a., „Nanobodies derived from Camelids represent versatile biomolecules for biomedical applications“, Biomater. Sci., Bd. 8, Nr. 13, S. 3559–3573, 2020, doi: 10.1039/D0BM00574F Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[16] J. Charles A Janeway, P. Travers, M. Walport, und M. J. Shlomchik, „The structure of a typical antibody molecule“, Immunobiol. Immune Syst. Health Dis. 5th Ed., 2001, Zugegriffen: Okt. 27, 2020. [Online]. Verfügbar unter: https://www.ncbi.nlm.nih.gov/books/NBK27144/.
[17] S. Muyldermans, „Nanobodies: natural single-domain antibodies“, Annu. Rev. Biochem., Bd. 82, S. 775–797, 2013, doi: 10.1146/annurev-biochem-063011-092449 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[18] „Nanobodies: Chemical Functionalization Strategies and Intracellular Applications - Schumacher - 2018 - Angewandte Chemie International Edition - Wiley Online Library“. https://onlinelibrary.wiley.com/doi/full/10.1002/anie.201708459 Titel anhand dieser DOI in Citavi-Projekt übernehmen (zugegriffen Okt. 27, 2020).
[19] D. Saerens, „Isolation and optimization of camelid single-domain antibodies: Dirk Saerens’ work on nanobodies“, World J. Biol. Chem., Bd. 1, Nr. 7, S. 235–238, Juli 2010, doi: 10.4331/wjbc.v1.i7.235 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[20] F. Rahbarizadeh, D. Ahmadvand, und Z. Sharifzadeh, „Nanobody; an Old Concept and New Vehicle for Immunotargeting“, Immunol. Invest., Bd. 40, Nr. 3, S. 299–338, Jan. 2011, doi: 10.3109/08820139.2010.542228 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[21] „Anwendungsgebiete für rekombinante Antikörper | SpringerLink“. https://link.springer.com/chapter/10.1007/978-3-662-50276-1_5 Titel anhand dieser DOI in Citavi-Projekt übernehmen (zugegriffen Okt. 27, 2020).
[22] M. Arbabi Ghahroudi, A. Desmyter, L. Wyns, R. Hamers, und S. Muyldermans, „Selection and identification of single domain antibody fragments from camel heavy-chain antibodies“, FEBS Lett., Bd. 414, Nr. 3, S. 521–526, Sep. 1997, doi: 10.1016/s0014-5793(97)01062-4 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[23] S. Schoonooghe u. a., „Novel applications of nanobodies for in vivo bio-imaging of inflamed tissues in inflammatory diseases and cancer“, Immunobiology, Bd. 217, Nr. 12, S. 1266–1272, Dez. 2012, doi: 10.1016/j.imbio.2012.07.009 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[24] „The Therapeutic Potential of Nanobodies | SpringerLink“. https://link.springer.com/article/10.1007/s40259-019-00392-z Titel anhand dieser DOI in Citavi-Projekt übernehmen (zugegriffen Okt. 27, 2020).
[25] S. Ewert, A. Honegger, und A. Plückthun, „Stability improvement of antibodies for extracellular and intracellular applications: CDR grafting to stable frameworks and structure-based framework engineering“, Methods San Diego Calif, Bd. 34, Nr. 2, S. 184–199, Okt. 2004, doi: 10.1016/j.ymeth.2004.04.007 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[26] M. Dondelinger u. a., „Understanding the Significance and Implications of Antibody Numbering and Antigen-Binding Surface/Residue Definition“, Front. Immunol., Bd. 9, Okt. 2018, doi: 10.3389/fimmu.2018.02278 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[27] „UCSF ChimeraX Home Page“. https://www.cgl.ucsf.edu/chimerax/ (zugegriffen Okt. 27, 2020).
[28] R. C. Cadwell und G. F. Joyce, „Randomization of genes by PCR mutagenesis“, PCR Methods Appl., Bd. 2, Nr. 1, S. 28–33, Aug. 1992, doi: 10.1101/gr.2.1.28 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[29] E. O. McCullum, B. A. R. Williams, J. Zhang, und J. C. Chaput, „Random mutagenesis by error-prone PCR“, Methods Mol. Biol. Clifton NJ, Bd. 634, S. 103–109, 2010, doi: 10.1007/978-1-60761-652-8_7 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[30] G. P. Smith, „Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface“, Science, Bd. 228, Nr. 4705, S. 1315–1317, Juni 1985, doi: 10.1126/science.4001944 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[31] J. McCafferty, A. D. Griffiths, G. Winter, und D. J. Chiswell, „Phage antibodies: filamentous phage displaying antibody variable domains“, Nature, Bd. 348, Nr. 6301, Art. Nr. 6301, Dez. 1990, doi: 10.1038/348552a0 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[32] A. N. Zacher, C. A. Stock, J. W. Golden, und G. P. Smith, „A new filamentous phage cloning vector: fd-tet“, Gene, Bd. 9, Nr. 1, S. 127–140, Apr. 1980, doi: 10.1016/0378-1119(80)90171-7 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[33] M. Better, C. P. Chang, R. R. Robinson, und A. H. Horwitz, „Escherichia coli secretion of an active chimeric antibody fragment“, Science, Bd. 240, Nr. 4855, S. 1041–1043, Mai 1988, doi: 10.1126/science.3285471 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[34] A. Skerra und A. Plückthun, „Assembly of a functional immunoglobulin Fv fragment in Escherichia coli“, Science, Bd. 240, Nr. 4855, S. 1038–1041, Mai 1988, doi: 10.1126/science.3285470 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[35] S. Bass, R. Greene, und J. A. Wells, „Hormone phage: an enrichment method for variant proteins with altered binding properties“, Proteins, Bd. 8, Nr. 4, S. 309–314, 1990, doi: 10.1002/prot.340080405 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[36] L. J. Garrard, M. Yang, M. P. O’Connell, R. F. Kelley, und D. J. Henner, „F AB Assembly and Enrichment in a Monovalent Phage Display System“, Bio/Technology, Bd. 9, Nr. 12, Art. Nr. 12, Dez. 1991, doi: 10.1038/nbt1291-1373 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[37] H. R. Hoogenboom, A. D. Griffiths, K. S. Johnson, D. J. Chiswell, P. Hudson, und G. Winter, „Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains.“, Nucleic Acids Res., Bd. 19, Nr. 15, S. 4133–4137, Aug. 1991.
[38] L. Ledsgaard, M. Kilstrup, A. Karatt-Vellatt, J. McCafferty, und A. H. Laustsen, „Basics of Antibody Phage Display Technology“, Toxins, Bd. 10, Nr. 6, Juni 2018, doi: 10.3390/toxins10060236 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[39] „Repertoires of aggregation-resistant human antibody domains | Protein Engineering, Design and Selection | Oxford Academic“. https://academic.oup.com/peds/article/20/8/413/1435168 (zugegriffen Okt. 27, 2020).
[40] C. F. Barbas, A. S. Kang, R. A. Lerner, und S. J. Benkovic, „Assembly of combinatorial antibody libraries on phage surfaces: the gene III site.“, Proc. Natl. Acad. Sci. U. S. A., Bd. 88, Nr. 18, S. 7978–7982, Sep. 1991.
[41] H. B. Lowman, S. H. Bass, N. Simpson, und J. A. Wells, „Selecting high-affinity binding proteins by monovalent phage display“, Biochemistry, Bd. 30, Nr. 45, S. 10832–10838, Nov. 1991, doi: 10.1021/bi00109a004 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[42] D. O’Connell, B. Becerril, A. Roy-Burman, M. Daws, und J. D. Marks, „Phage versus Phagemid Libraries for Generation of Human Monoclonal Antibodies“, J. Mol. Biol., Bd. 321, Nr. 1, S. 49–56, Aug. 2002, doi: 10.1016/S0022-2836(02)00561-2 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[43] U. Brinkmann, P. S. Chowdhury, D. M. Roscoe, und I. Pastan, „Phage display of disulfide-stabilized Fv fragments“, J. Immunol. Methods, Bd. 182, Nr. 1, S. 41–50, Jan. 1995, doi: 10.1016/0022-1759(95)00016-4 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[44] J. Yan, G. Li, Y. Hu, W. Ou, und Y. Wan, „Construction of a synthetic phage-displayed Nanobody library with CDR3 regions randomized by trinucleotide cassettes for diagnostic applications“, J. Transl. Med., Bd. 12, Nr. 1, S. 343, Dez. 2014, doi: 10.1186/s12967-014-0343-6 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[45] A. D. Griffiths u. a., „Isolation of high affinity human antibodies directly from large synthetic repertoires.“, EMBO J., Bd. 13, Nr. 14, S. 3245–3260, Juli 1994.
[46] T. Clackson, H. R. Hoogenboom, A. D. Griffiths, und G. Winter, „Making antibody fragments using phage display libraries“, Nature, Bd. 352, Nr. 6336, S. 624–628, Aug. 1991, doi: 10.1038/352624a0 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[47] J. D. Marks, H. R. Hoogenboom, T. P. Bonnert, J. McCafferty, A. D. Griffiths, und G. Winter, „By-passing immunization: Human antibodies from V-gene libraries displayed on phage“, J. Mol. Biol., Bd. 222, Nr. 3, S. 581–597, Dez. 1991, doi: 10.1016/0022-2836(91)90498-U Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[48] R. E. Hawkins, S. J. Russell, und G. Winter, „Selection of phage antibodies by binding affinity: Mimicking affinity maturation“, J. Mol. Biol., Bd. 226, Nr. 3, S. 889–896, Aug. 1992, doi: 10.1016/0022-2836(92)90639-2 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[49] S. R. Whaley, D. S. English, E. L. Hu, P. F. Barbara, und A. M. Belcher, „Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly“, Nature, Bd. 405, Nr. 6787, S. 665–668, Juni 2000, doi: 10.1038/35015043 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[50] M. Dondelinger u. a., „Understanding the Significance and Implications of Antibody Numbering and Antigen-Binding Surface/Residue Definition“, Front. Immunol., Bd. 9, 2018, doi: 10.3389/fimmu.2018.02278 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[51] A. Honegger und A. Plückthun, „Yet Another Numbering Scheme for Immunoglobulin Variable Domains: An Automatic Modeling and Analysis Tool“, J. Mol. Biol., Bd. 309, Nr. 3, S. 657–670, Juni 2001, doi: 10.1006/jmbi.2001.4662 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[52] R. C. Cadwell und G. F. Joyce, „Randomization of genes by PCR mutagenesis“, PCR Methods Appl., Bd. 2, Nr. 1, S. 28–33, Aug. 1992, doi: 10.1101/gr.2.1.28 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[53] E. O. McCullum, B. A. R. Williams, J. Zhang, und J. C. Chaput, „Random mutagenesis by error-prone PCR“, Methods Mol. Biol. Clifton NJ, Bd. 634, S. 103–109, 2010, doi: 10.1007/978-1-60761-652-8_7 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[54] N. E. Biolabs, „OneTaq® DNA Polymerase | NEB“. https://international.neb.com/products/m0480-onetaq-dna-polymerase (zugegriffen Okt. 28, 2020).
[55] N. E. Biolabs, „Hot Start Taq DNA Polymerase | NEB“. https://international.neb.com/products/m0495-hot-start-taq-dna-polymerase (zugegriffen Okt. 28, 2020).
[56] N. E. Biolabs, „Q5® High-Fidelity DNA Polymerase | NEB“. https://international.neb.com/products/m0491-q5-high-fidelity-dna-polymerase (zugegriffen Okt. 28, 2020).
[57] J. B. GmbH, „JBS Error-Prone Kit, Molecular Evolution: Random Mutagenesis“, Jena Bioscience. https://www.jenabioscience.com/molecular-biology/cloning-and-mutagenesis/random-mutagenesis-kits/pp-102-jbs-error-prone-kit (zugegriffen Okt. 28, 2020).
[58] „Quality control NGS libraries, Bioanalyzer High Sensitivity DNA | Agilent“. https://www.agilent.com/en/product/automated-electrophoresis/bioanalyzer-systems/bioanalyzer-dna-kits-reagents/bioanalyzer-high-sensitivity-dna-analysis-228249#productdetails (zugegriffen Okt. 28, 2020).
[59] „igvtools | Integrative Genomics Viewer“. https://software.broadinstitute.org/software/igv/igvtools (zugegriffen Okt. 28, 2020).
[60] „Understanding Illumina Quality Scores“, S. 2.
[61] N. E. Biolabs, „SfiI | NEB“. https://international.neb.com/products/r0123-sfii (zugegriffen Okt. 28, 2020).
[62] N. E. Biolabs, „Impact | NEB“. https://international.neb.com/products/protein-expression-and-purification-technologies/e-coli/impact/impact (zugegriffen Okt. 28, 2020).
[63] „Engineered Recombinant Single-Chain Fragment Variable Antibody for Immunosensors | Analytical Chemistry“. https://pubs.acs.org/doi/pdf/10.1021/ac0507690 Titel anhand dieser DOI in Citavi-Projekt übernehmen (zugegriffen Okt. 28, 2020).
[64] R. P. D. Bank, „1D PFV: 1JNH“. https://www.rcsb.org/sequence/1JNH (zugegriffen Okt. 28, 2020).
[65] R. P. D. Bank, „1D PFV: 1DBB“. https://www.rcsb.org/sequence/1DBB (zugegriffen Okt. 28, 2020).
[66] „geneart-seamless-cloning-assembly-mutagenesis-tools-brochure.pdf“. Zugegriffen: Okt. 28, 2020. [Online]. Verfügbar unter: https://tools.thermofisher.com/content/sfs/brochures/geneart-seamless-cloning-assembly-mutagenesis-tools-brochure.pdf.
[67] Z. Shen, G. A. Stryker, R. L. Mernaugh, L. Yu, H. Yan, und X. Zeng, „Single-Chain Fragment Variable Antibody Piezoimmunosensors“, Anal. Chem., Bd. 77, Nr. 3, S. 797–805, Feb. 2005, doi: 10.1021/ac048655w Titel anhand dieser DOI in Citavi-Projekt übernehmen.
[68] A. M. Azevedo, V. C. Martins, D. M. Prazeres, V. Vojinović, J. M. Cabral, und L. P. Fonseca, „Horseradish peroxidase: a valuable tool in biotechnology“, Biotechnol. Annu. Rev., Bd. 9, S. 199–247, 2003, doi: 10.1016/s1387-2656(03)09003-3 Titel anhand dieser DOI in Citavi-Projekt übernehmen.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]


