Difference between revisions of "Part:BBa K3136004"
| Line 4: | Line 4: | ||
It is a conding sequence of CRISPR-Cas12a from Francisella novicida U112. The sequence is coden optimized. | It is a conding sequence of CRISPR-Cas12a from Francisella novicida U112. The sequence is coden optimized. | ||
| + | |||
| + | =Improvement= | ||
| + | Sequence Analysis | ||
| + | We blast the sequences of our new part BBa_K3521001 and the existing part BBa_K3136004. As shown in the figure, there is 2852bp inconsistent good, 1058bp inconsistent bad from the old one part. | ||
| + | [[File:T--SHSID-BBa_K3521001 Fig 6.jpg|600px|thumb|center|Figure 7 The Blast Results of BBa_K3521001 and BBa_K3136004]] | ||
| + | |||
| + | Different Function | ||
| + | First of all, we aim to detect different organisms for different illnesses. The old part is used to detect the African Swine fever virus (ASFV). Our new part is used to detect the presence of Listeria monocytogenes, which is easily survival in low temperature like refrigerator environment, and then cause Listeriosis illness, like fever, muscle ache, and so on. | ||
| + | |||
| + | Furthermore, the old part only designed a crRNA that just targeting a site of ASFV, which may have less wide application. In contrast, we design six different crRNAs that can guide our improved Cas12a (FnCpf1) protein targeting six different genomic sites of Listeria monocytogenes, so that our new part can detect more extensive Listeria monocytogenes. | ||
| + | -------------------------------------------------------------------------------------------------- | ||
| + | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
Revision as of 19:47, 27 October 2020
FnCas12a-typeIIS
It is a conding sequence of CRISPR-Cas12a from Francisella novicida U112. The sequence is coden optimized.
Improvement
Sequence Analysis We blast the sequences of our new part BBa_K3521001 and the existing part BBa_K3136004. As shown in the figure, there is 2852bp inconsistent good, 1058bp inconsistent bad from the old one part.
Different Function First of all, we aim to detect different organisms for different illnesses. The old part is used to detect the African Swine fever virus (ASFV). Our new part is used to detect the presence of Listeria monocytogenes, which is easily survival in low temperature like refrigerator environment, and then cause Listeriosis illness, like fever, muscle ache, and so on.
Furthermore, the old part only designed a crRNA that just targeting a site of ASFV, which may have less wide application. In contrast, we design six different crRNAs that can guide our improved Cas12a (FnCpf1) protein targeting six different genomic sites of Listeria monocytogenes, so that our new part can detect more extensive Listeria monocytogenes.
Usage and Biology
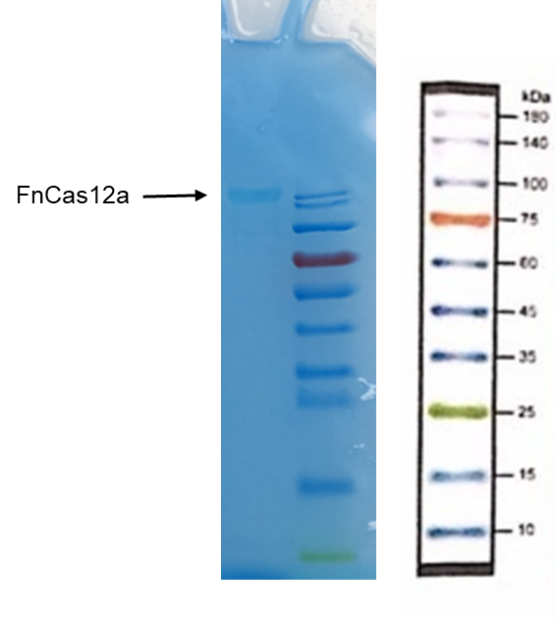
BBa_K3136004 FnCas12a-typeIIS
Methods:
1. Put the EP tube containing purified FnCas12a protein at 99℃ water for 15 minutes.
2. After heating, centrifuge the tubes for 5min.
3. Take and install the gel, add the electrophoresis buffer to the sample hole, and check if there is any leakage. Add marker, to the first and last sample holes, add FnCas12a to the other sample holes, respectively.
7. Connect the electrophoresis device to the power supply. Connect the positive electrode to the tank and the negative electrode to the slot for electrophoresis. The voltage is adjusted to 160V.
8. Turn off the power supply and disconnect the electrode until the bromophenol blue reaches the bottom of the release adhesive. Remove the glass sheet from the electrophoresis device and then remove the gel.
9. Soak the gel in Coomassie brilliant blue dye and dye it in a horizontal shaking bed for 15 min.
Observe the protein bands
Cas12a detection method:
Materials:
Water. 14.5 μL
Buffer. 2 μL
Enzyme (Cas12a). 0.5 μL
Template. 0.5 μL
(From solution we made after LAMP)
Detector. 2 μL
crRNA. 0.5 μL
(Note: the Deteror is sequence of ssDNA HEX-N12-BHQ1 or FAM-N12-BHQ1 or FITC-T14-Biotin)
Procedure:
1. Add all the materials together:
• Firstly, use pipette to straw 14.5 μL pure water from the pure water bottle to one tube.
• Secondly, use pipette to straw 2 μL buffer to the previous tube.
• Thirdly, use pipette to straw 0.5 μL enzyme to the previous tube.
• Next, use pipette to straw 0.5 μL template to the previous tube.
• Then, add 0.5 μL crRNA to the previous tube.
• Finally, use pipette to straw 2 μL detector to the previous tube.
2. Straw 1 μL of the mixed solution in the tubes to the 96-well Plate.
3. Detect the solution using fluorescent detector machine to check the fluorescence when they react 10min.
Results:
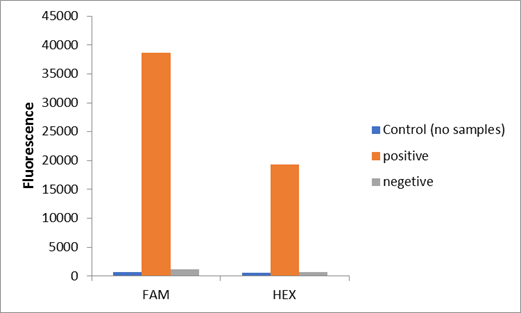
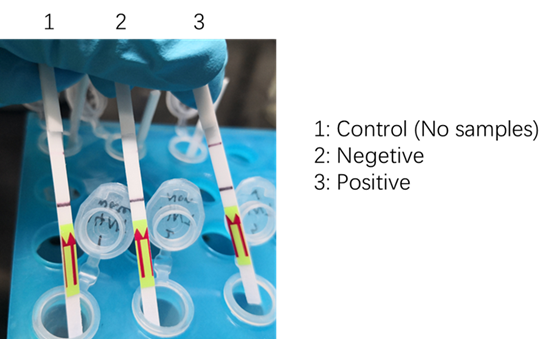
We used two types of fluorescent probe (FAM and HEX) to check the fluorescence of FnCas12a reaction system at 10min as shown in Fig1. Linked to the control and negative samples, the positive sample has a high fluorescence. It means that FnCas12a slices the fluorescent probe in the reaction system and the cracked fluorescent probe produce fluorescence. we also checked the fluorescence of FnCas12a reaction system at 10min by lateral flow using briDetect (Milenia Biotec GmbH) (Fig2).
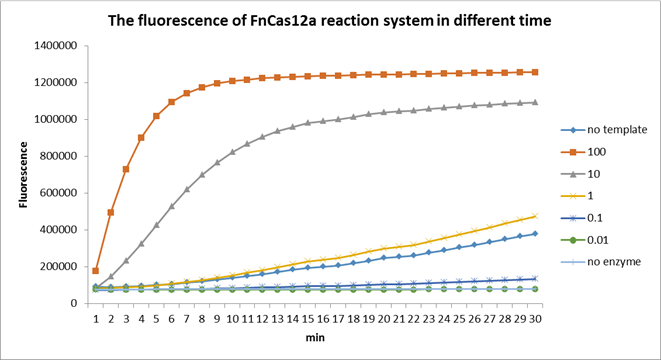
Using fluorescent detector machine, we can detect the fluorescence of FnCas12a reaction system at different time by setting up program properly in the machine. We detect the fluorescence under different concentration FnCas12a and different time(Fig3). With the time growth, the fluorescence gradually raised until a certain time. Higher concentration FnCas12a had higher fluorescence and the fluorescence increased quickly.
Reference
Li S Y, Cheng Q X, Liu J K, et al. CRISPR-Cas12a has both cis-and trans-cleavage activities on single-stranded DNA[J]. Cell research, 2018, 28(4): 491.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 310
Illegal PstI site found at 364
Illegal PstI site found at 1627
Illegal PstI site found at 2533
Illegal PstI site found at 3727 - 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 310
Illegal PstI site found at 364
Illegal PstI site found at 1627
Illegal PstI site found at 2533
Illegal PstI site found at 3727 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1253
Illegal BglII site found at 1586
Illegal BglII site found at 1706
Illegal BglII site found at 2000
Illegal BglII site found at 2444
Illegal BglII site found at 3335
Illegal BglII site found at 3425 - 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 310
Illegal PstI site found at 364
Illegal PstI site found at 1627
Illegal PstI site found at 2533
Illegal PstI site found at 3727 - 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 310
Illegal PstI site found at 364
Illegal PstI site found at 1627
Illegal PstI site found at 2533
Illegal PstI site found at 3727
Illegal NgoMIV site found at 2480
Illegal NgoMIV site found at 3319
Illegal NgoMIV site found at 3530 - 1000COMPATIBLE WITH RFC[1000]